It looks like you're using an Ad Blocker.
Please white-list or disable AboveTopSecret.com in your ad-blocking tool.
Thank you.
Some features of ATS will be disabled while you continue to use an ad-blocker.
share:
a reply to: soficrow
Well, now that is a lot of info, info that make my spidey senses tingle, anyways, very very interesting...
Wouldn't at one point I guess this whole bio computer, figure out how to create life? Not sure if that makes sense, but if there is biology involved, wouldn't it get there? Kinda like we did....hrmm....will need to come back for this with more time...but thank you!
Well, now that is a lot of info, info that make my spidey senses tingle, anyways, very very interesting...
Wouldn't at one point I guess this whole bio computer, figure out how to create life? Not sure if that makes sense, but if there is biology involved, wouldn't it get there? Kinda like we did....hrmm....will need to come back for this with more time...but thank you!
a reply to: eldemie
Would it figure out how to create life by itself? Not really. No. The processes that appear to be responsible for making life are fairly complicated and vary quite a bit. We only know a fraction of them and many of the ones we know about appear to be more like evolutionary accidents.
Even defining life gets a bit complicated, as people vary the definition. Is a plant alive? A fungus? A virus?
Interestingly, viruses are at the edge of non-life and life. They are not alive because they don't have metabolism. They steal from organisms that do have metabolism--meaning they can't convert energy into cellular activity, nor offspring. So they're like machines, missing some important parts, but with the ability to replicate by stealing the energy and materials of living cells.
Let's look at science's attempt to develop simple life:
Source: motherboard.vice.com...
This number is important (437 genes). Genes are sections of DNA or RNA that carry information that was passed down to the organism. At some point in the past, basic genetic machinery was passing down genetic information without actually being alive. Scientists believe this was possible because of areas (perhaps all around Earth) that were rich in resources and heat--where the materials could naturally merge together and slowly develop into more complex forms. This process wouldn't be much different than mixing ingredients together for dinner, then putting them in the oven. Although, this process would take many more cycles in and out of the oven.
To create an optimized, simple working organism, it took 437 selected genes. For nature to create a successful working organism, it would most likely require much large number of genes, far more than 437, because many genes wouldn't directly help the organism. That's the downside of trial and error. There is a lot more error than success. It can also require several generations for successful mutations to be expressed in future generations (epigenetics), but that's another topic.
All of these aspects have to be created/developed: "capacity to grow, metabolize, respond (to stimuli), adapt, and reproduce".
1. Metabolism appears to be the hardest.
2. Many chemical compounds can grow, or interconnect with themselves or complementary units. But I think growth regulation would also be a critical part.
3. Responding to stimuli is also a natural part of chemicals interacting, so that's an easier one to explain.
4. Adaptations would slowly accumulate in the "primordial soup", where these various components would start to accumulate in the same pre-organism.
5. Replicating genes is not quite as difficult, assuming you have the component parts. Correcting errors in gene replication is difficult, but putting that aside for now, just pairing up genes in a complementary strand is not incredibly complicated.
Ultimately, metabolism is needed to be a self-contained organism--meaning you can replicate your own parts without needed the help of an outside source. You only need something that you can convert into fuel and basic "nutrients" or "chemicals" from the environment--this is "food"... energy and parts for replication and repair.
So, why do I say "no"? The problem really comes from the amount of time it takes for the development of random mutations and turning them into a viable organism, without using a template. A computer could recreate an organism from a template, but I don't think it's going to accidentally build an organism from scratch, as the number of random evolutions needed would require an incredibly long time (longer than the life of the computer).
SciShow created some videos about the creation of life on Earth. Life was able to achieve a primordial cell relatively quickly (maybe in the first billion years), but it took billions of years before it could overcome its limitations:
It wouldn't figure it out on its own. We could program it to look for the particular mechanisms that we use for life, but the computer wouldn't be developing life by itself. It would just be running a program to select for our particular form of life.
For completeness, here is an article on NASA scientists developing 3 of the 4 bases needed for DNA/RNA in a lab setting:
www.space.com...
Would it figure out how to create life by itself? Not really. No. The processes that appear to be responsible for making life are fairly complicated and vary quite a bit. We only know a fraction of them and many of the ones we know about appear to be more like evolutionary accidents.
Even defining life gets a bit complicated, as people vary the definition. Is a plant alive? A fungus? A virus?
Life - A distinctive characteristic of a living organism from dead organism or non-living thing, as specifically distinguished by the capacity to grow, metabolize, respond (to stimuli), adapt, and reproduce.
Interestingly, viruses are at the edge of non-life and life. They are not alive because they don't have metabolism. They steal from organisms that do have metabolism--meaning they can't convert energy into cellular activity, nor offspring. So they're like machines, missing some important parts, but with the ability to replicate by stealing the energy and materials of living cells.
Let's look at science's attempt to develop simple life:
Source: motherboard.vice.com...
a self-replicating bacterium invented by Venter and his team that contains just 437 genes, a "genome smaller than that of any autonomously replicating cell found in nature"
This number is important (437 genes). Genes are sections of DNA or RNA that carry information that was passed down to the organism. At some point in the past, basic genetic machinery was passing down genetic information without actually being alive. Scientists believe this was possible because of areas (perhaps all around Earth) that were rich in resources and heat--where the materials could naturally merge together and slowly develop into more complex forms. This process wouldn't be much different than mixing ingredients together for dinner, then putting them in the oven. Although, this process would take many more cycles in and out of the oven.
To create an optimized, simple working organism, it took 437 selected genes. For nature to create a successful working organism, it would most likely require much large number of genes, far more than 437, because many genes wouldn't directly help the organism. That's the downside of trial and error. There is a lot more error than success. It can also require several generations for successful mutations to be expressed in future generations (epigenetics), but that's another topic.
All of these aspects have to be created/developed: "capacity to grow, metabolize, respond (to stimuli), adapt, and reproduce".
1. Metabolism appears to be the hardest.
2. Many chemical compounds can grow, or interconnect with themselves or complementary units. But I think growth regulation would also be a critical part.
3. Responding to stimuli is also a natural part of chemicals interacting, so that's an easier one to explain.
4. Adaptations would slowly accumulate in the "primordial soup", where these various components would start to accumulate in the same pre-organism.
5. Replicating genes is not quite as difficult, assuming you have the component parts. Correcting errors in gene replication is difficult, but putting that aside for now, just pairing up genes in a complementary strand is not incredibly complicated.
Ultimately, metabolism is needed to be a self-contained organism--meaning you can replicate your own parts without needed the help of an outside source. You only need something that you can convert into fuel and basic "nutrients" or "chemicals" from the environment--this is "food"... energy and parts for replication and repair.
So, why do I say "no"? The problem really comes from the amount of time it takes for the development of random mutations and turning them into a viable organism, without using a template. A computer could recreate an organism from a template, but I don't think it's going to accidentally build an organism from scratch, as the number of random evolutions needed would require an incredibly long time (longer than the life of the computer).
SciShow created some videos about the creation of life on Earth. Life was able to achieve a primordial cell relatively quickly (maybe in the first billion years), but it took billions of years before it could overcome its limitations:
Wouldn't at one point I guess this whole bio computer, figure out how to create life?
It wouldn't figure it out on its own. We could program it to look for the particular mechanisms that we use for life, but the computer wouldn't be developing life by itself. It would just be running a program to select for our particular form of life.
For completeness, here is an article on NASA scientists developing 3 of the 4 bases needed for DNA/RNA in a lab setting:
www.space.com...
a reply to: soficrow
In the last minute of this interview (starting at 6:08), Anne Condon, the head of computer science at UBC explains that in about 50 years (so the year 2065) she expects that molecular programming and DNA programming will be more commonplace in medicine.
I thought this was worth posting, as she leads research in the area of bioinformatics.
She talks, more in detail, about molecular programming in this talk:
In the last minute of this interview (starting at 6:08), Anne Condon, the head of computer science at UBC explains that in about 50 years (so the year 2065) she expects that molecular programming and DNA programming will be more commonplace in medicine.
I thought this was worth posting, as she leads research in the area of bioinformatics.
She talks, more in detail, about molecular programming in this talk:
a reply to: Protector
What if we were to thrown some quantum mechanics in there? Would that change the outcome? With the subject about it changes when we see it...when we don't etc? You're making me double up on the coffee haha!
Hmm, will need more time then, but keeping this close, gracias!
What if we were to thrown some quantum mechanics in there? Would that change the outcome? With the subject about it changes when we see it...when we don't etc? You're making me double up on the coffee haha!
Hmm, will need more time then, but keeping this close, gracias!
originally posted by: soficrow
a reply to: Protector
Nice post. Thanks. But...
You forgot to consider prions.
...the computer ...would just be running a program to select for our particular form of life.
A dangerous assumption.
I'm glad you enjoyed it.
I have extremely limited knowledge of prions. I haven't run across them, yet, in bioinformatics. I don't really see where they'd be great in molecular computing. There are many other ways to manipulate molecules where you don't have to simultaneously worry about infection. Let's face it... almost no one wants to work in a biohazard environment.
Biohazard levels for prions are BSL-2 or BSL-3: www.cureffi.org...
Do you really want to work in this suit all the time?
en.wikipedia.org...#/media/File:Influenza_virus_research.jpg
Not fun.
Interestingly, I just found this article on DNA computing, published yesterday...
Source: www.csmonitor.com...
Currently, the cost and time required for this process is somewhat prohibitive for consumer applications. It cost $7,000 to synthesize the DNA Erlich developed and another $2,000 to read it. The synthesis process took two weeks and the sequencing took about a day.
It confirms my suspicions about prohibitive costs.
Regarding a computer that selects for a particular form of life:
Remember that computers just do what they're instructed to do. Almost no one wants to be the one responsible for eating up super computer time on a program that never finishes. It's just not realistic. Those things are expensive... like, super duper expensive. Computer scientists do a lot of planning, as well as a lot of trial and error while developing a program. A program that would need to "create life" would have a daunting task. Realistically, it'd need limitations so that it could actually accomplish something.
So, I don't really think it's a dangerous assumption. I'm the type of guy that they'd ask to write such a thing. If we could throw "practicality" out of the window, I can open up some options. But we can't throw it out of the window. Come back to me when Quantum Computing is ready and we might be able to take some awesome shortcuts that make this exercise much more feasible... maybe.
a reply to: eldemie
Quantum computing can overcome problems in something called BQP space. That's the formal name of problems that can be solved easily by a quantum computer. However, since that probably sounds like nonsense, I'll have to refer you to yet another video.
Keep your eye out for BQP... (this may require more coffee)
Oh, and if you meant something else by "quantum mechanics", please explain.
Quantum computing can overcome problems in something called BQP space. That's the formal name of problems that can be solved easily by a quantum computer. However, since that probably sounds like nonsense, I'll have to refer you to yet another video.
Keep your eye out for BQP... (this may require more coffee)
Oh, and if you meant something else by "quantum mechanics", please explain.
a reply to: Protector
Hmm. Proteins are where it's at in bioinformatics and synthetic biology. The synthetic biology market is huge - predicted to top $5.6 Billion by 2018. Cell-free protein synthesis (CFPS) is a large part of that market.
Prion proteins are incredibly stable, certainly more stable than ordinary proteins and DNA, and the obvious choice for use in protein-based bio-computing. True, the documentation and coverage -for biotech's CFPS and McGill's "Living Computer"- do not describe their proteins as prions. However they would have to be, considering the inherent instability of regular proteins and DNA.
Prions are infectious, yes, but only on contact with proteins of a similar kind. Given that industry is working with non-natural synthetic proteins, this characteristic would prevent 'cross-species' infectivity, theoretically at least. As well, the infectious capacity implies superior abilities for self-repair and memory retrieval. [Prions store memories.]
I have extremely limited knowledge of prions. I haven't run across them, yet, in bioinformatics. I don't really see where they'd be great in molecular computing. There are many other ways to manipulate molecules where you don't have to simultaneously worry about infection. Let's face it... almost no one wants to work in a biohazard environment.
Hmm. Proteins are where it's at in bioinformatics and synthetic biology. The synthetic biology market is huge - predicted to top $5.6 Billion by 2018. Cell-free protein synthesis (CFPS) is a large part of that market.
Prion proteins are incredibly stable, certainly more stable than ordinary proteins and DNA, and the obvious choice for use in protein-based bio-computing. True, the documentation and coverage -for biotech's CFPS and McGill's "Living Computer"- do not describe their proteins as prions. However they would have to be, considering the inherent instability of regular proteins and DNA.
Prions are infectious, yes, but only on contact with proteins of a similar kind. Given that industry is working with non-natural synthetic proteins, this characteristic would prevent 'cross-species' infectivity, theoretically at least. As well, the infectious capacity implies superior abilities for self-repair and memory retrieval. [Prions store memories.]
Synthetic Biology Market worth $5,630.4 Million by 2018
[$5.6+ Billion]
Cell-free protein synthesis (also called in-vitro protein synthesis or abbreviated CFPS), is the production of protein using biological machinery without the use of living cells.
The three major end-users of CFPS systems are the pharmaceutical & biotechnological industry, contract research institutes, and academic & research institutes, of which pharmaceutical companies are dominant, followed by contract research institutes.
The living supercomputer
...HOW IS THE 'LIVING COMPUTER' MORE EFFICIENT?
* The agents (proteins) are available in large numbers at negligible cost.
* They are self-propelled and thus do not require a global, external driving force;
* Proteins operate independently of each other to ensure parallel exploration and have small dimensions to enable use in high-density networks with high computing power per unit area.
Building living, breathing supercomputers
2012. The lure of molecular computing: While marketable products seem decades away, researchers are crystallizing theories and devices that will give biological organisms the power to compute
Pico-world dragnets: Computer-designed proteins recognize and bind small molecules
Computer-designed proteins that can recognize and interact with small biological molecules are now a reality. ...
“This is major step toward building proteins for use as biosensors or molecular sponges, or in synthetic biology — giving organisms new tools to perform a task,” said one of the lead researchers, Christine E. Tinberg, a postdoctoral fellow in biochemistry at the UW.
Protein Structure, Modelling and Applications
1. Why Is It Important to Study Proteins?
In the drama of life on a molecular scale, proteins are where the action is (1).
Proteins are molecular devices, in the nanometer scale, where biological function is exerted (1). They are the building blocks of all cells in our bodies and in all living creatures of all kingdoms. Although the information necessary for life to go on is encoded by the DNA molecule, the dynamic process of life maintenance, replication, defense and reproduction are carried out by proteins.
...[If... then...] the target protein 3D structure can be modeled based on one or on a combination of several template molecules (26). This is possible because homolog proteins descend from a common ancestor and are likely to present the same structure and function. Caution note: This is correct in general, but there are exceptions though. [sic]
edit on 3/3/17 by soficrow because: (no reason given)
edit on 3/3/17 by soficrow because: (no reason given)
a reply to: soficrow
You're right that proteins are used all the time. Proteins are what create everything, as they are directly translated from RNA, which is transcribed from our DNA. Proteins and DNA are the bread and butter of bioinformatics. But Prions are a very specific thing.
A $5.6 billion market isn't that big in terms of the $3+ trillion healthcare industry in the U.S. As I've come to find out recently, the healthcare industry is absolutely massive and far-reaching. There are hundreds of specializations, each doing research into different areas.
www.forbes.com...
You can't really compare protein research, or even most synthetic biology research, with Prions. The really hot topic right now is CRISPR/Cas9. I was talking with a Biology Ph.D. (specializing in mammalian reproduction) about it recently. Theoretically, it should unlock a lot of avenues for advancing targeted genetic manipulation.
But my only understanding of the molecular computing industry is that it is possibly several dozen years away from becoming a reality. Graphene technology was supposed to be one of the first big keys in creating efficient molecules. But it is actually behind schedule. Mass producing high quality graphene has been quite difficult and expensive. I feel (although I may be wrong) that graphene is a bit of a litmus test for the industry. If an area with thousands of researchers, billions of dollars in funding, and huge industrial applications is having trouble bringing a new generation of molecular materials to life, I don't have high hopes that other areas of the industry will fly past them. Does that make sense?
In short, I wouldn't bet that Prions are a major part of upcoming molecular computing, nor protein research. It's more like a fringe technology sitting in the corner of a much larger and more well funded marketplace. But that's mostly speculation.
You're right that proteins are used all the time. Proteins are what create everything, as they are directly translated from RNA, which is transcribed from our DNA. Proteins and DNA are the bread and butter of bioinformatics. But Prions are a very specific thing.
A $5.6 billion market isn't that big in terms of the $3+ trillion healthcare industry in the U.S. As I've come to find out recently, the healthcare industry is absolutely massive and far-reaching. There are hundreds of specializations, each doing research into different areas.
www.forbes.com...
You can't really compare protein research, or even most synthetic biology research, with Prions. The really hot topic right now is CRISPR/Cas9. I was talking with a Biology Ph.D. (specializing in mammalian reproduction) about it recently. Theoretically, it should unlock a lot of avenues for advancing targeted genetic manipulation.
But my only understanding of the molecular computing industry is that it is possibly several dozen years away from becoming a reality. Graphene technology was supposed to be one of the first big keys in creating efficient molecules. But it is actually behind schedule. Mass producing high quality graphene has been quite difficult and expensive. I feel (although I may be wrong) that graphene is a bit of a litmus test for the industry. If an area with thousands of researchers, billions of dollars in funding, and huge industrial applications is having trouble bringing a new generation of molecular materials to life, I don't have high hopes that other areas of the industry will fly past them. Does that make sense?
In short, I wouldn't bet that Prions are a major part of upcoming molecular computing, nor protein research. It's more like a fringe technology sitting in the corner of a much larger and more well funded marketplace. But that's mostly speculation.
a reply to: Protector
I'm not comparing - I'm saying prion creation is inevitable whether accidentally or purposefully. Moreover, prions are incredibly stable whereas normal proteins and DNA are incredibly unstable - so prions are the obvious choice for use in biocomputing.
[But yes, CRISPR is hot too.]
Again, prions are the most stable bio-component, by far. Moreover, proteins misfold at the drop of a hat - and misfolded proteins can become prions, even when that is not the intention. [I just hope they're not flushing all their failures into the sewage system like Big Pharma used to do.]
...You can't really compare protein research, or even most synthetic biology research, with Prions.
I'm not comparing - I'm saying prion creation is inevitable whether accidentally or purposefully. Moreover, prions are incredibly stable whereas normal proteins and DNA are incredibly unstable - so prions are the obvious choice for use in biocomputing.
[But yes, CRISPR is hot too.]
In short, I wouldn't bet that Prions are a major part of upcoming molecular computing, nor protein research. It's more like a fringe technology sitting in the corner of a much larger and more well funded marketplace. But that's mostly speculation.
Again, prions are the most stable bio-component, by far. Moreover, proteins misfold at the drop of a hat - and misfolded proteins can become prions, even when that is not the intention. [I just hope they're not flushing all their failures into the sewage system like Big Pharma used to do.]
a reply to: soficrow
DNA is incredibly stable. It's a crystalline form (made from sugars). It takes significant temperature to disrupt it and quickly repairs damage (within the body).
Proteins do have a lot of weak spots. They can be transcribed or translated incorrectly. They can be denatured in a variety of ways. They can fold incorrectly. They can be deactivated depending on their receptors and the environment they are exposed to. But that relates to why they can do almost anything. They're amazingly flexible. If DNA/RNA comprise the white and black keys of a piano, the codons would be the chords, and the proteins would be the music.
If I come across anything Prion related in the near future, I can revisit this thread.
DNA is incredibly stable. It's a crystalline form (made from sugars). It takes significant temperature to disrupt it and quickly repairs damage (within the body).
Proteins do have a lot of weak spots. They can be transcribed or translated incorrectly. They can be denatured in a variety of ways. They can fold incorrectly. They can be deactivated depending on their receptors and the environment they are exposed to. But that relates to why they can do almost anything. They're amazingly flexible. If DNA/RNA comprise the white and black keys of a piano, the codons would be the chords, and the proteins would be the music.
If I come across anything Prion related in the near future, I can revisit this thread.
originally posted by: Protector
a reply to: soficrow
DNA is incredibly stable.
Humph.
DNA samples stored at 4°C and RT showed varying degrees of evaporation but DNA was stable for up to 12 months at 4°C. Samples stored at room temperature totally evaporated by 6 months (Figure 2). At RT, DNA degradation was seen at 9 months. DNA stored in dry state at room temperature showed degradation at 3 months of storage (Figure 4).
Proteins do have a lot of weak spots. They can be transcribed or translated incorrectly. They can be denatured in a variety of ways. They can fold incorrectly. They can be deactivated depending on their receptors and the environment they are exposed to.
2xhumph. Yes, and when they do, they can become stable prions.
...prion polypeptides are intrinsically predisposed to non-physiological folding conformations that would account for their environmental durability
It is a commonplace observation that the prion forms of proteins are stable...
There are ...numerous other synthetic prions, consisting of sequences derived from those of prion-forming proteins modified by deletion, mutagenesis or by fusion with heterologous natural or artificial sequences for functional modification or as reporters.
But that relates to why they can do almost anything. They're amazingly flexible.
lol. Like I said. When they become prions.
If I come across anything Prion related in the near future, I can revisit this thread.
Absolutely. Feel free.
edit on 3/3/17 by soficrow because: (no reason given)
a reply to: soficrow
I still think you're over-applying prion formation out of proteins. Proteins do not commonly denature into prions. Specific proteins found in the brain seem to convert into prions at an advanced age. And, from what it seems, there is probably a genetic component to the formation of prions in a lot of people, as the diseases associated with them are largely inherited.
But I'll keep in mind your love for prions. I have been reading some Alzheimer's research, lately, so maybe I'll see them pop up in there.
And I still think DNA is incredibly stable. I mean, it lasts for months and it's only a handful of atoms across. Sure, diamond's carbon structure has it beat, but I'm still impressed by DNA.
I still think you're over-applying prion formation out of proteins. Proteins do not commonly denature into prions. Specific proteins found in the brain seem to convert into prions at an advanced age. And, from what it seems, there is probably a genetic component to the formation of prions in a lot of people, as the diseases associated with them are largely inherited.
But I'll keep in mind your love for prions. I have been reading some Alzheimer's research, lately, so maybe I'll see them pop up in there.
And I still think DNA is incredibly stable. I mean, it lasts for months and it's only a handful of atoms across. Sure, diamond's carbon structure has it beat, but I'm still impressed by DNA.
originally posted by: Protector
a reply to: soficrow
I still think you're over-applying prion formation out of proteins. Proteins do not commonly denature into prions. Specific proteins found in the brain seem to convert into prions at an advanced age.
I'm using the original definition of prion - "proteinaceous infectious particle." In fact, virtually any protein can misfold and become a prion.
Your information comes from a "communications strategy" initiated with the "Mad Cow" disease scare. The "Mad Cow"-affected protein in the human brain was renamed "the prion protein," and the disease agent called a "prion." This effectively obscured other infectious mis-folded proteins, confusing the public and many scientists working outside the field, and deflected attention from the issue. Which was, of course, the intent.
Note though, it appears that few prions cause disease - and in the main, serve as an incredibly rapid and effective adaptation device in the evolutionary scheme of things.
To clarify further, when I talk about prions I am not referring to THE one-and-only-Mad-Cow-prion strain. I am talking about prions in general - proteins that have mis-folded to become infectious prions.
And I still think DNA is incredibly stable. I mean, it lasts for months and it's only a handful of atoms across. Sure, diamond's carbon structure has it beat, but I'm still impressed by DNA.
...DNA lasts for months. Not at all useful for building bio-computers, methinks. Prions, on the other hand, have been conserved over hundreds of millions of years. However, given that our world's waters and soil have been rather well-contaminated by nasty suckers like CWD, and not to mention taken up by plants and grasses, I can see why this little info tidbit might be subject to "scientific-information-censorship" too.
]
DNA Dethroned - Inheritance is Protein-Based.
...prions -and the traits they confer- can be inherited;
in humans, some are conserved over hundreds of millions of years.
]
Prion-like Protein Discovered in Bacteria
[…the emergence of prions predates the evolutionary split between eukaryotes and bacteria - and DNA]
edit on 4/3/17 by soficrow because: (no reason given)
a reply to: soficrow
It appears that you have a possible factual error:
By that logic, DNA has been conserved for something like 5 billion years.
The actual quote is (source: Your Link):
That means that human DNA (and human ancestor DNA) has conserved the genetic encoding for proteins that become prions.
I assume (correct me if I'm wrong) that you believe the prions are being physically passed down because of this statement:
That's in relation to the epigenetic variations between cells. Prions aren't inherited separately from DNA. I don't know if you assumed they were or not.
What scientists are testing has to do with an oddity they found in yeast.
Source: Protein based inheritance
So they don't always know why, when we have 2 versions of the protein, the epigenetic markers enable/activate the prion version of the protein (i.e. the disease version).
Let me add another quote:
The prion version can have an environmental advantage for yeast. It does beg the question, "What advantage do they have in humans that makes it so we would inherit them?" Although that might be a bad question, because the amyloid plaques from Alzheimer's may be a common protein folding error from degradation, not necessarily a separate version of the protein.
I believe I read a paper a while back stating that the plaques in the brain of Alzheimer's patients might be part of an immune response.
Here is something along those lines: Alzheimers disease, prions, and the immune system
Anyway, prions are not more resilient than DNA. They are created from DNA.
Here is a link on epigenetic inheritance.
It appears that you have a possible factual error:
DNA lasts for months. Not at all useful for building bio-computers, methinks. Prions, on the other hand, have been conserved over hundreds of millions of years.
By that logic, DNA has been conserved for something like 5 billion years.
The actual quote is (source: Your Link):
When the team examined the human cognates (i.e. blood relatives) of the prion-proteins, the intrinsically disordered domains were conserved over hundreds of millions of years.
That means that human DNA (and human ancestor DNA) has conserved the genetic encoding for proteins that become prions.
I assume (correct me if I'm wrong) that you believe the prions are being physically passed down because of this statement:
A team of Whitehead Institute and Stanford University scientists are redefining what it means to be a prion--a type of protein that can pass heritable traits from cell to cell by its structure instead of by DNA.
That's in relation to the epigenetic variations between cells. Prions aren't inherited separately from DNA. I don't know if you assumed they were or not.
What scientists are testing has to do with an oddity they found in yeast.
Source: Protein based inheritance
This property suggested a basis for inheritance of the altered protein. In a normal yeast life cycle the Sup35p stays soluble. However, in a [PSI+] strain the prion form aggregates. Lindquist describes this as a "renegade protein whose misfolding creates a surface that other proteins of the same type will add on to." The altered proteins can also be transferred from a mother to a daughter cell. Lindquist's group tied the in vivo process to the in vitro process by using mutations that affected the inheritance of the prions and also affected the protein's ability to form amyloids in vitro.
...
Though researchers have a simple biochemical model for prion formation, they still don't know the underlying mechanism of prion inheritance. The protein forms amyloid rich in b structures, and new proteins join on to it, but the driving force behind the molecular changes is a mystery.
So they don't always know why, when we have 2 versions of the protein, the epigenetic markers enable/activate the prion version of the protein (i.e. the disease version).
Let me add another quote:
"Protein-based genetic elements allow cells to have two different heritable phenotypic (physical) states with the same genome," comments Linquist. For example, it is biologically advantageous for cells to have [PSI] (the prion form) under some circumstances. A large yeast colony is likely to have one of these altered proteins ready to exploit a change in environment. On the other hand, if the environment doesn't change, the cell can drop out of the colony. "It is an ancient mechanism of inheritance, but a newly appreciated one," she adds.
The prion version can have an environmental advantage for yeast. It does beg the question, "What advantage do they have in humans that makes it so we would inherit them?" Although that might be a bad question, because the amyloid plaques from Alzheimer's may be a common protein folding error from degradation, not necessarily a separate version of the protein.
I believe I read a paper a while back stating that the plaques in the brain of Alzheimer's patients might be part of an immune response.
Here is something along those lines: Alzheimers disease, prions, and the immune system
Anyway, prions are not more resilient than DNA. They are created from DNA.
Here is a link on epigenetic inheritance.
edit on 2017-3-4 by Protector because: Fixing a possible confusing part of my response
edit on 2017-3-4 by Protector because:
Adding a link and fixing broken links
a reply to: soficrow
Soficrow,
I am going to say that it likely got zapped by that EO mentioned.
But hey, at least the links and initial articles still EXIST on the internet, heck they're even still on their original sites!
In the early 2000's I ran across an interesting piece on some research going on across MULTIPLE universities including searchable databases with academic papers freely available.
Myself and 3 other people archived big chunks of data and the original article, including burning backups to Cd-rom.
The article: gone not even on way back
The papers: Same
The university websites: Same
OK so that's enough to make someone a bit shaky.
It gets worse!
Everyone's archives on their net connected PC's and sometimes connected laptops:
GONE
The CD-ROM's
Gone or unrecoverably corrupted!
If it had happened to just me or I was the only witness, I would write the whole thing off.
It didn't just happen to me!
I've had two other incidents of a similar nature since then.
It seems like intellectual curiosity is far from considered a virtue these days.
Soficrow,
I am going to say that it likely got zapped by that EO mentioned.
But hey, at least the links and initial articles still EXIST on the internet, heck they're even still on their original sites!
In the early 2000's I ran across an interesting piece on some research going on across MULTIPLE universities including searchable databases with academic papers freely available.
Myself and 3 other people archived big chunks of data and the original article, including burning backups to Cd-rom.
The article: gone not even on way back
The papers: Same
The university websites: Same
OK so that's enough to make someone a bit shaky.
It gets worse!
Everyone's archives on their net connected PC's and sometimes connected laptops:
GONE
The CD-ROM's
Gone or unrecoverably corrupted!
If it had happened to just me or I was the only witness, I would write the whole thing off.
It didn't just happen to me!
I've had two other incidents of a similar nature since then.
It seems like intellectual curiosity is far from considered a virtue these days.
I take the pragmatic approach on this.
I am with those who think bio computing was dropped when the concept of quantum computing began to take shape.
There's no point spending time and money developing a slower "next gen" computer when you can likely spend the same on the ultimate computer.
I am with those who think bio computing was dropped when the concept of quantum computing began to take shape.
There's no point spending time and money developing a slower "next gen" computer when you can likely spend the same on the ultimate computer.
originally posted by: Protector
a reply to: soficrow
...human DNA (and human ancestor DNA) has conserved the genetic encoding for proteins that become prions.
No. DNA codes for proteins - prions are considered part of epigenetic inheritance, which does NOT change DNA.
I assume (correct me if I'm wrong) that you believe the prions are being physically passed down because of this statement:
No, not one single statement. I've been monitoring prion research for over a decade - the evidence showing prion heritability is extensive and solid.
That's in relation to the epigenetic variations between cells. Prions aren't inherited separately from DNA. I don't know if you assumed they were or not.
You are the one making assumptions based on old science and communications strategies.
Anyway, prions are not more resilient than DNA. They are created from DNA.
Prions certainly are FAR more resilient than DNA. And while a few may be coded for, they are not all "created from DNA." Far from it. And from your source:
Epigenetic inheritance is an unconventional finding. It goes against the idea that inheritance happens only through the DNA code that passes from parent to offspring. It means that a parent's experiences, in the form of epigenetic tags, can be passed down to future generations.
As unconventional as it may be, there is little doubt that epigenetic inheritance is real.
edit on 5/3/17 by soficrow because: (no reason given)
edit on 5/3/17 by soficrow because: format
edit on
5/3/17 by soficrow because: (no reason given)
a reply to: roguetechie
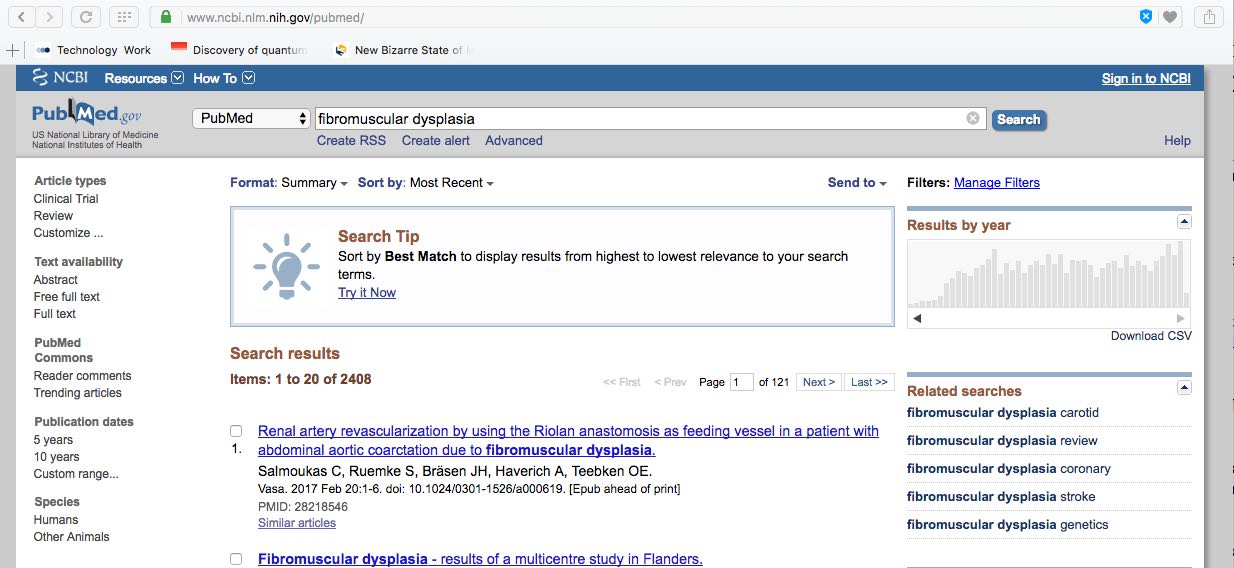
I had the same experience in the early 2000's while researching a disease called "fibromuscular dysplasia," which is recognized officially as causing cell changes to smooth muscles in blood vessel walls.
PubMed listed over 1.5 million articles for a "fibromuscular dysplasia" (FMD) search starting in the late 1990's when I began my project. Much of the data had to do with FMD cell changes occurring in different organs and systems, and its appearance in different diseases.
By the early 2000's, I had started playing with the hypothesis that the myofibroblasts generated in FMD resulted from a prion related to the actin protein. I posted my research and ideas on the internet for discussion.
One morning (December 4, 2003?) while I was on PubMed searching and reading FMD related articles (still with over 1.5 million on the list), I flipped from an article back to my search to find a messages saying, "This topic is now closed."
I tried several times to get the PubMed list back, failed. Then I realized that the US censorship would likely spread, so I scooted over to the other medical databases from other countries I had on file, madly searching, copying and grabbing another research article. ...They closed down one after the other, and displayed the same message: "This topic is now closed."
Like others, I raised a huge stink on the Internet about "censoring medical and scientific information." After a few weeks, PubMed announced the topic was "closed for review" and would open in 3 months. When it did open, the available research articles were culled back to about 50. Even now, over 10 years later, a PubMed search pulls up less than 3000 papers.
In the early 2000's I ran across an interesting piece on some research going on across MULTIPLE universities including searchable databases with academic papers freely available.
Myself and 3 other people archived big chunks of data and the original article, including burning backups to Cd-rom.
The article: gone not even on way back
The papers: Same
The university websites: Same
OK so that's enough to make someone a bit shaky.
It gets worse!
I had the same experience in the early 2000's while researching a disease called "fibromuscular dysplasia," which is recognized officially as causing cell changes to smooth muscles in blood vessel walls.
PubMed listed over 1.5 million articles for a "fibromuscular dysplasia" (FMD) search starting in the late 1990's when I began my project. Much of the data had to do with FMD cell changes occurring in different organs and systems, and its appearance in different diseases.
By the early 2000's, I had started playing with the hypothesis that the myofibroblasts generated in FMD resulted from a prion related to the actin protein. I posted my research and ideas on the internet for discussion.
One morning (December 4, 2003?) while I was on PubMed searching and reading FMD related articles (still with over 1.5 million on the list), I flipped from an article back to my search to find a messages saying, "This topic is now closed."
I tried several times to get the PubMed list back, failed. Then I realized that the US censorship would likely spread, so I scooted over to the other medical databases from other countries I had on file, madly searching, copying and grabbing another research article. ...They closed down one after the other, and displayed the same message: "This topic is now closed."
Like others, I raised a huge stink on the Internet about "censoring medical and scientific information." After a few weeks, PubMed announced the topic was "closed for review" and would open in 3 months. When it did open, the available research articles were culled back to about 50. Even now, over 10 years later, a PubMed search pulls up less than 3000 papers.

new topics
-
German city in chaos as 'extremist' march sees calls for 'caliphate' and ISIS-style flags
Mainstream News: 1 hours ago -
College protesters want amnesty.
US Political Madness: 2 hours ago -
Shocking moment four men 'try to force Jewish pedestrian into car boot' in North London
Breaking Alternative News: 5 hours ago -
The 'Censorship-Industrial Complex'. It is coming to a nation state near you, any time now...
New World Order: 6 hours ago -
BREAKING: Astrazeneca admits for the first time its vaccine can cause deaths and serious injuries
Medical Issues & Conspiracies: 10 hours ago
top topics
-
New Bombshell Evidence Strongly Suggests Trump was Set Up in Classified Docs Saga
US Political Madness: 15 hours ago, 36 flags -
BREAKING: Astrazeneca admits for the first time its vaccine can cause deaths and serious injuries
Medical Issues & Conspiracies: 10 hours ago, 14 flags -
Shocking moment four men 'try to force Jewish pedestrian into car boot' in North London
Breaking Alternative News: 5 hours ago, 9 flags -
German city in chaos as 'extremist' march sees calls for 'caliphate' and ISIS-style flags
Mainstream News: 1 hours ago, 8 flags -
One More Night at the Pig and Blanket (Time 2024)
Short Stories: 17 hours ago, 6 flags -
College protesters want amnesty.
US Political Madness: 2 hours ago, 4 flags -
The 'Censorship-Industrial Complex'. It is coming to a nation state near you, any time now...
New World Order: 6 hours ago, 1 flags
active topics
-
German city in chaos as 'extremist' march sees calls for 'caliphate' and ISIS-style flags
Mainstream News • 4 • : nugget1 -
College protesters want amnesty.
US Political Madness • 14 • : dothedew -
Expert Says Parents Should Ask Babies Permission to Change Nappies.
General Chit Chat • 46 • : FlyersFan -
New Bombshell Evidence Strongly Suggests Trump was Set Up in Classified Docs Saga
US Political Madness • 50 • : xuenchen -
Shocking moment four men 'try to force Jewish pedestrian into car boot' in North London
Breaking Alternative News • 37 • : DerBeobachter2 -
University of Texas Instantly Shuts Down Anti Israel Protests
Education and Media • 395 • : FlyersFan -
New whistleblower Jason Sands speaks on Twitter Spaces last night.
Aliens and UFOs • 84 • : gippo88 -
Hate makes for strange bedfellows
US Political Madness • 66 • : 19Bones79 -
Candidate TRUMP Now Has Crazy Judge JUAN MERCHAN After Him - The Stormy Daniels Hush-Money Case.
Political Conspiracies • 819 • : Oldcarpy2 -
Gov Kristi Noem Shot and Killed "Less Than Worthless Dog" and a 'Smelly Goat
2024 Elections • 135 • : SideEyeEverything1
