It looks like you're using an Ad Blocker.
Please white-list or disable AboveTopSecret.com in your ad-blocking tool.
Thank you.
Some features of ATS will be disabled while you continue to use an ad-blocker.
share:
a reply to: Xtrozero
We also have a very modern examples of mutation with the animals of Chernobyl
Interesting article here, dogs are dogs, cats are cats and horses are horses.
Chernobyl Animal Facts
A side point about all these animals returning to the area was this; essentially, this means that human populations have a bigger negative impact than radiation, pretty sad when you think about it.
We also have a very modern examples of mutation with the animals of Chernobyl
Interesting article here, dogs are dogs, cats are cats and horses are horses.
Chernobyl Animal Facts
A side point about all these animals returning to the area was this; essentially, this means that human populations have a bigger negative impact than radiation, pretty sad when you think about it.
originally posted by: Blue_Jay33
originally posted by: Barcs
originally posted by: cooperton
originally posted by: Barcs
It happens alongside evolution, as I clearly said.
Epigenetic inheritance is repeatable in a lab. Evolution is not. There has never been an organism to change into another organism over time via the mechanisms of evolution. Your statement is faith-based.
Another 100% lie. Seriously give up the ghost, Kent. You are wasting everyone's time with your unconvincing verbal diarrhea and ignorant diatribe.
Funny how yet again, you ignore the vast majority of the post, in favor of a dishonest quote mine. JUST LIKE ALWAYS.
Lab based empirical evidence with proven science verses your faith based theory....now who is lying ?
Have you actually studied this yourself or are you simply parroting Cooperton?
This is the problem when you don’t actually understand the material enough to properly discuss it aside from a bunch of non sequitors and “rah rah go team Hovind” posts. I’m not sure which is actually worse though. Having read the material and coming to the same conclusion as Coop or enjoying the bliss of willful ignorance and simply repeating what you want the truth to be.
I’ll say one thing. Cooperton is correct that Epigenetic Inheritance has been show to occur in the lab. But only in unicellular organisms. From cell to cell. Not generationally from parent to child. See, there’s actually a big difference between Epigenetic Inheritance and Transgenerational Epigenetic Inheritance. Cooperton is trying to claim the latter is equivalent to the former when this has NOT been shown in a lab.
So please... cite the “Lab based empirical evidence with proven science” that supports your nonsense.
edit on 25-7-2019 by peter vlar
because: (no reason given)
originally posted by: Noinden
a reply to: cooperton
Actually thanks to Next Gen DNA sequencing, you can repeat any number of experiments, as long as you have a PC with enough memory, and a bit of patience. You do need to know what you are looking at, but anyone, and I mean ANYONE can check results.
DNA studies have shown evolution to be correct.
Fanciful science fiction like this, I always enjoy it.
a reply to: Blue_Jay33
It really must suck for you, when your premise is proven wrong.
You seem to think empiricism is only achieved one way. Which amply illustrates that you are just throwing the word around, with out an understanding of it.
It really must suck for you, when your premise is proven wrong.
You seem to think empiricism is only achieved one way. Which amply illustrates that you are just throwing the word around, with out an understanding of it.
a reply to: Noinden
Concluding paragraph of that link.
"Cognitive Bias" in science.
Concluding paragraph of that link.
researchers need to be more assertive that evolution has both occurred, and continues to occur. It is essential that any person who does not accept the continuity of evolution puts forward alternative testable models. As we tell our first year undergraduates, ‘belief is the curse of the thinking class’.
"Cognitive Bias" in science.
originally posted by: Blue_Jay33
a reply to: Noinden
Concluding paragraph of that link.
researchers need to be more assertive that evolution has both occurred, and continues to occur. It is essential that any person who does not accept the continuity of evolution puts forward alternative testable models. As we tell our first year undergraduates, ‘belief is the curse of the thinking class’.
"Cognitive Bias" in science.
Not even close to cognitive bias. That would be on your end. You somehow ignored the second sentence you cited. It was -
It is essential that any person who does not accept the continuity of evolution puts forward alternative testable models.
That’s not any type of bias at all. What that is saying, is that you’re free to believe whatever you like. But, and this part is important, the onus is on you to support it with a testable model. Not once has any proponent of any version of Creationism demonstrated a testable model of anything. They haven’t even claimed to have had a model and then flaked on showing it. They simply don’t have one other than “Duh.... obviously god did it because I can’t wrap my head around it all enough to understand any other possibilities”. Which is the default response to anything that is a little out of their reach.
a reply to: peter vlar
Then you are free to feel as the report says "belief is the curse of the thinking class" and that is a prejudice about a possibility and thus a cognitive bias, in particular something called selective perception.
Then you are free to feel as the report says "belief is the curse of the thinking class" and that is a prejudice about a possibility and thus a cognitive bias, in particular something called selective perception.
originally posted by: cooperton
originally posted by: Phantom423
a reply to: cooperton
More garbage. You pick and choose whatever you think validates your point.
It doesn't.
Garbage.
No no no, I don't pick and choose, you chose to bring up microtubules. I am simply showing, from your link, why it requires other factors to be in play, and therefore does not 'self-assemble' without the guidance of the rest of necessary proteins and catalysts present within the cell. So this is another example of irreducible complexity due to the reasons said above.
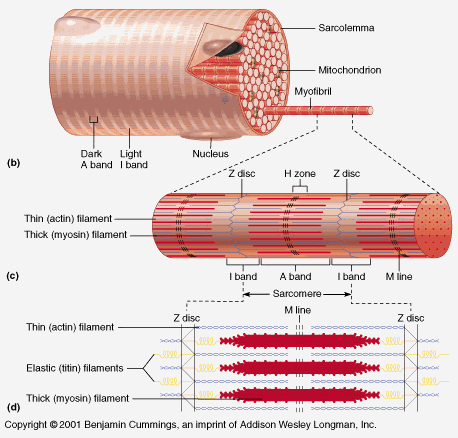
Literally every protein component in the body exhibits this same dependence on other components to exist. You also brought up the 'self-assembly' of myosin. The paper you presented was from 1972 and you didn't even have access to it. More recent research no longer calls it 'self-assembly', instead they more accurately call it 'assembly'. Here is their description of myosin formation:
"Myofibrillogenesis in striated muscle cells requires a precise ordered pathway to assemble different proteins into a linear array of sarcomeres. The sarcomere relies on interdigitated thick and thin filaments to ensure muscle contraction, as well as properly folded and catalytically active myosin head. Achieving this organization requires a series of protein folding and assembly steps. The folding of the myosin head domain requires chaperone activity to attain its functional conformation. Folded or unfolded myosin can spontaneously assemble into short myosin filaments, but further assembly requires the short and incomplete myosin filaments to assemble into the developing thick filament. These longer filaments are then incorporated into the developing sarcomere of the muscle. Both myosin folding and assembly require factors to coordinate the formation of the thick filament in the sarcomere and these factors include chaperone molecules. Myosin folding and sarcomeric assembly requires association of classical chaperones as well as folding cofactors such as UNC-45. Recent research has suggested that UNC-45 is required beyond initial myosin head folding and may be directly or indirectly involved in different stages of myosin thick filament assembly, maintenance and degradation."
Source
Notice how in the quote above they discuss the multitude of steps and other proteins involved in forming one of the many proteins involved in muscle formation. This shows it could not have arisen by piece-by-piece sequential mutations via evolution, because it requires all necessary components to function.
You'll have to clarify a few things:
You also brought up the 'self-assembly' of myosin. The paper you presented was from 1972 and you didn't even have access to it.
Which paper are you referring to that I don't have access to? Are you referring to the paper that the video is based on?
Notice how in the quote above they discuss the multitude of steps and other proteins involved in forming one of the many proteins involved in muscle formation. This shows it could not have arisen by piece-by-piece sequential mutations via evolution, because it requires all necessary components to function.
I think you may have shot yourself in the foot on this one. Of course there are a multitude of steps involved in the process - any process for that matter. That's what self assembly is about. It's a process of assembly.
What exactly do you want to discuss with regard to self assembly? If it's about a specific paper, please post the paper you're referring to. If it's the process of self assembly itself, then let's start a new thread which focuses exclusively on self assembly.
I agree with the paper: the process of myosin assembly involves "a precise ordered pathway to assemble different proteins into a linear array of sarcomeres (muscle units)". When protein chains are created, they need to be folded properly to function.
This goes to the thermodynamics of self assembly. Similar to the post where I suggested the kitchen experiment, this is not a mystery. Self assembly is a thermodynamic process. Protein folding is a classic example - that's exactly what I said in a previous post.
So again, please describe your problem with the process of self assembly. I think your vision or interpretation of what self assembly is leaves out a lot of detail.
originally posted by: Phantom423
Of course there are a multitude of steps involved in the process - any process for that matter. That's what self assembly is about. It's a process of assembly.
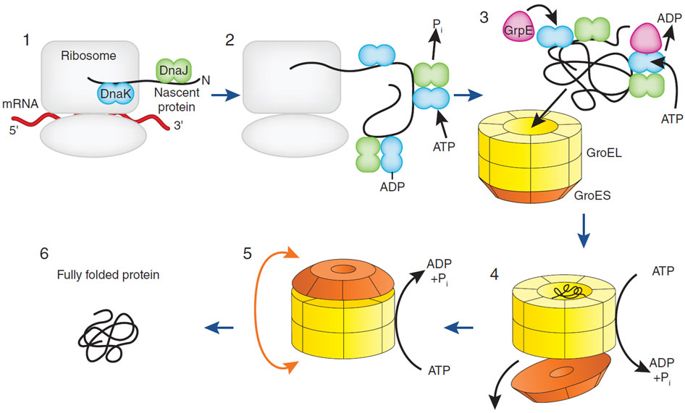
Self-assembly by definition would be something assembling by itself without external influences. This would allow it to by-pass the dilemma of irreducible complexity. But, protein sequences do not self assemble, especially myosin. They need transcription, translation, and then post-translational modifications, such as the chaperone folding proteins discussed in the paper: myosin assembly
This goes to the thermodynamics of self assembly. Self assembly is a thermodynamic process. Protein folding is a classic example - that's exactly what I said in a previous post.
But by definition it's not self assembly. If it were self-assembly the proteins would be able to fold on their own, but they require chaperone proteins:
Since myosin cannot self assemble, nor can the other components necessary to create a muscle unit, then it remains irreducibly complex. This is why sequential modifications by evolution would not have been able to create such a synchronized unit.
originally posted by: peter vlar
(epigenetic inheritance has only been shown) in unicellular organisms. From cell to cell. Not generationally from parent to child. See, there’s actually a big difference between Epigenetic Inheritance and Transgenerational Epigenetic Inheritance. Cooperton is trying to claim the latter is equivalent to the former when this has NOT been shown in a lab.
So please... cite the “Lab based empirical evidence with proven science” that supports your nonsense.
It has been shown in mammals too.
"However, some recent animal studies suggest an apparent resistance to complete erasure of epigenetic marks during early development, enabling transgenerational epigenetic inheritance. " Source
It was once dogma that the epigenome had to be erased to allow proper cell differentiation in a developing embryo (i.e. blank slate stem cells), but now they're finding that there are ways that the epigenome is inherited from the parents.
" it is now clear that inheritance not based on DNA sequence exists in multiple organisms, with examples found in microbes, plants, and invertebrate and vertebrate animals."
Source 2
a reply to: cooperton
Exactly what "external influences" are you referring to? Self assembly is a PROCESS. Of course there are many steps in the process. What would you expect?
So what is it that you object to with self assembly? Protein sequences do self assemble.
Self-organization: the fundament of cell biology
Roland Wedlich-Söldner and Timo Betz
Published:09 April 2018 doi.org...
____________________________________________________________________
Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus
Diana Born, Lukas Reuter, Ulrike Mersdorf, Melanie Mueller, Matthias G. Fischer, Anton Meinhart, and Jochen Reinstein
PNAS July 10, 2018 115 (28) 7332-7337; first published June 25, 2018
doi.org...
Edited by Michael G. Rossmann, Purdue University, West Lafayette, IN, and approved May 30, 2018 (received for review March 28, 2018)
So what is it that you don't understand about self assembly? If it's that confusing, go to the methodology and see how these experiments were done.
If self assembly wasn't part of biological systems, there would be no cell regeneration and no life.
Self-assembly by definition would be something assembling by itself without external influences. This would allow it to by-pass the dilemma of irreducible complexity. But, protein sequences do not self assemble, especially myosin. They need transcription, translation, and then post-translational modifications, such as the chaperone folding proteins discussed in the paper: myosin assembly
Exactly what "external influences" are you referring to? Self assembly is a PROCESS. Of course there are many steps in the process. What would you expect?
So what is it that you object to with self assembly? Protein sequences do self assemble.
Self-organization: the fundament of cell biology
Roland Wedlich-Söldner and Timo Betz
Published:09 April 2018 doi.org...
Abstract
Self-organization refers to the emergence of an overall order in time and space of a given system that results from the collective interactions of its individual components. This concept has been widely recognized as a core principle in pattern formation for multi-component systems of the physical, chemical and biological world. It can be distinguished from self-assembly by the constant input of energy required to maintain order—and self-organization therefore typically occurs in non-equilibrium or dissipative systems. Cells, with their constant energy consumption and myriads of local interactions between distinct proteins, lipids, carbohydrates and nucleic acids, represent the perfect playground for self-organization. It therefore comes as no surprise that many properties and features of self-organized systems, such as spontaneous formation of patterns, nonlinear coupling of reactions, bi-stable switches, waves and oscillations, are found in all aspects of modern cell biology.
Ultimately, self-organization lies at the heart of the robustness and adaptability found in cellular and organismal organization, and hence constitutes a fundamental basis for natural selection and evolution.
____________________________________________________________________
Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus
Diana Born, Lukas Reuter, Ulrike Mersdorf, Melanie Mueller, Matthias G. Fischer, Anton Meinhart, and Jochen Reinstein
PNAS July 10, 2018 115 (28) 7332-7337; first published June 25, 2018
doi.org...
Edited by Michael G. Rossmann, Purdue University, West Lafayette, IN, and approved May 30, 2018 (received for review March 28, 2018)
Abstract
Virophages have the unique property of parasitizing giant viruses within unicellular hosts. Little is understood about how they form infectious virions in this tripartite interplay. We provide mechanistic insights into assembly and maturation of mavirus, a marine virophage, by combining structural and stability studies on capsomers, virus-like particles (VLPs), and native virions.
We found that the mavirus protease processes the double jelly-roll (DJR) major capsid protein (MCP) at multiple C-terminal sites and that these sites are conserved among virophages. Mavirus MCP assembled in Escherichia coli in the absence and presence of penton protein, forming VLPs with defined size and shape. While quantifying VLPs in E. coli lysates, we found that full-length rather than processed MCP is the competent state for capsid assembly. Full-length MCP was thermally more labile than truncated MCP, and crystal structures of both states indicate that full-length MCP has an expanded DJR core.
Thus, we propose that the MCP C-terminal domain serves as a scaffolding domain by adding strain on MCP to confer assembly competence. Mavirus protease processed MCP more efficiently after capsid assembly, which provides a regulation mechanism for timing capsid maturation. By analogy to Sputnik and adenovirus, we propose that MCP processing renders mavirus particles infection competent by loosening interactions between genome and capsid shell and destabilizing pentons for genome release into host cells.
The high structural similarity of mavirus and Sputnik capsid proteins together with conservation of protease and MCP processing suggest that assembly and maturation mechanisms described here are universal for virophages.
So what is it that you don't understand about self assembly? If it's that confusing, go to the methodology and see how these experiments were done.
If self assembly wasn't part of biological systems, there would be no cell regeneration and no life.
a reply to: cooperton
What do you mean "by definition"????? THIS IS A PROCESS. The chaperone proteins are part of the process. How do you think the chaperone proteins formed? The entire footprint is a self assembled process. Structure/function of any organism does not occur in a vacuum. IT IS A PROCESS. Please get that through your head once and for all.
But by definition it's not self assembly. If it were self-assembly the proteins would be able to fold on their own, but they require chaperone proteins:
What do you mean "by definition"????? THIS IS A PROCESS. The chaperone proteins are part of the process. How do you think the chaperone proteins formed? The entire footprint is a self assembled process. Structure/function of any organism does not occur in a vacuum. IT IS A PROCESS. Please get that through your head once and for all.
originally posted by: Blue_Jay33
originally posted by: Barcs
originally posted by: cooperton
originally posted by: Barcs
It happens alongside evolution, as I clearly said.
Epigenetic inheritance is repeatable in a lab. Evolution is not. There has never been an organism to change into another organism over time via the mechanisms of evolution. Your statement is faith-based.
Another 100% lie. Seriously give up the ghost, Kent. You are wasting everyone's time with your unconvincing verbal diarrhea and ignorant diatribe.
Funny how yet again, you ignore the vast majority of the post, in favor of a dishonest quote mine. JUST LIKE ALWAYS.
Lab based empirical evidence with proven science verses your faith based theory....now who is lying ?
en.wikipedia.org...
originally posted by: cooperton
Do you have any demonstrable evidence that an organism can change into another organism over time? Like real empirical lab data.
I've posted it dozens of times, and you ignorantly find weak excuses to ignore/dismiss it every single time. You've been given SO MUCH evidence by Phantom and others on here and you never ever address it. You'll take 1 line of the whole thing and jump to wild conclusions about it and make ignorant generalizations that have nothing to do with the research itself. It happens every time, without fail, just like with my epigenetics post. You completely ignored all of it and quickly diverted to the bull# lie that epigenetics is demonstrable in a lab while evolution is not. Sorry, it doesn't get any more dishonest than that.
edit on 7 26 19 by Barcs because: (no reason given)
a reply to: cooperton
There are 38 references included with this paper. This is the PHYSICS of protein folding. Pay attention for a change and learn something.
The physics of protein self-assembly
Author links open overlay panelJennifer J.McManusaPatrickCharbonneaubEmanuelaZaccarellicdNeerAsherieef
Show more
doi.org...

There are 38 references included with this paper. This is the PHYSICS of protein folding. Pay attention for a change and learn something.
The physics of protein self-assembly
Author links open overlay panelJennifer J.McManusaPatrickCharbonneaubEmanuelaZaccarellicdNeerAsherieef
Show more
doi.org...

Abstract
Understanding protein self-assembly is important for many biological and industrial processes. Proteins can self-assemble into crystals, filaments, gels, and other amorphous aggregates. The final forms include virus capsids and condensed phases associated with diseases such as amyloid fibrils. Although seemingly different, these assemblies all originate from fundamental protein interactions and are driven by similar thermodynamic and kinetic factors. Here we review recent advances in understanding protein self-assembly through a soft condensed matter perspective with an emphasis on three specific systems: globular proteins, viruses, and amyloid fibrils. We conclude with a discussion of unanswered questions in the field.
a reply to: cooperton
I've posted enough links which describe self assembly as a physiological process in nature. At the very least, you should be able to understand that self assembly is a well elucidated phenomenon.
If you think all these authors are wrong, please write them a letter.
The excerpt below is from a Biochemistry book.
Collagen
Volume I: Biochemistry
ByMarcel E. Nimni
Chapter 2
Energetics and Thermodynamics of Collagen Self-Assembly
With George Némethy
P.S. Please don't bother to tell me that you don't have access to the book.
I've posted enough links which describe self assembly as a physiological process in nature. At the very least, you should be able to understand that self assembly is a well elucidated phenomenon.
If you think all these authors are wrong, please write them a letter.
The excerpt below is from a Biochemistry book.
Collagen
Volume I: Biochemistry
ByMarcel E. Nimni
Chapter 2
Energetics and Thermodynamics of Collagen Self-Assembly
With George Némethy
The structure of collagen is characterized by the hierarchical order of self-assembly of its constituent units. Physicochemical methods, including thermodynamic measurements and considerations of energetics have been used to study the structure and conformation of the polypeptide chains, the formation and stability of the triple helix, and, to a lesser extent, the association of triple helices. This chapter deals with these levels of self-assembly mainly from the relationships between structure and the energetics of noncovalent interactions. The conformational properties of the polypeptide chains in collagen and the interactions that determine conformation and assembly are the same as those that govern the folding of globular proteins. Upon heating, collagen and collagen-like poly(tripeptide)s undergo a transition over a narrow temperature range. The enthalpy of melting of monodisperse solutions has been determined by calorimetry for many collagen species and for synthetic model analogs. The effect of salts and denaturants on collagen melting is similar to their effect on the denaturation of globular proteins.
P.S. Please don't bother to tell me that you don't have access to the book.
edit on 26-7-2019 by Phantom423 because: (no reason given)
a reply to: Phantom423
in response to this paper
It is not self-assembly in the sense that the protein sequence assembled by itself. That process still needs transcription and translation to parse the DNA code and make the polypeptide chain. There are no articles that show that the process of peptide polymerization can "self-assemble". So you aren't really resolving the problem of these proteins "self-assembling", because they still require the DNA code and the machinery thereafter to be made. Without transcription and translation, coherent polypeptide chains cannot be made in nature.
Your source is discussing the self-folding of polypeptide chains. So this isn't self-assembly like you think it is, they are explaining the various ways of folding these peptide chains into the tertiary structured proteins. They mention pathogenic folding, or in other words, undesirably folded proteins. This occurs when there is not proper post-translational folding. As per the example used in the paper you provided, amyloid fibrils are the pathogenic correlate of Alzheimer's disease. Another example they use is viral capsid formation. But this isn't necessarily self-assembly as you think, it still requires the host cell's machinery to forego transcription and translation to make the polypeptide sequence.
I do not doubt that tertiary and quaternary protein structures can self-assemble, in fact, biological organisms need chaperone proteins to literally chaperone the folding of the secondary protein structure (the polypeptide chain) into a coherent tertiary and quaternary structure. Failure to do so leads to all sorts of pathogenesis, i.e. amyloid fibrils.

So your wish that complex functional proteins can just self-assemble from scratch peptide monomers is totally unfounded in the literature. Transcription and translation are still required, and they can only parse sections of DNA that code for particular proteins... so the dilemma still remains, how could all of the proteins required for the muscle unit have been created by synchronous mutations? Especially considering the titin protein has over 100,000 base pairs (DNA units) of length to be coded for.
It is absolutely unfounded in the literature how such a synchronized event could have occurred to overcome the leap of irreducible complexity.
in response to this paper
It is not self-assembly in the sense that the protein sequence assembled by itself. That process still needs transcription and translation to parse the DNA code and make the polypeptide chain. There are no articles that show that the process of peptide polymerization can "self-assemble". So you aren't really resolving the problem of these proteins "self-assembling", because they still require the DNA code and the machinery thereafter to be made. Without transcription and translation, coherent polypeptide chains cannot be made in nature.
Your source is discussing the self-folding of polypeptide chains. So this isn't self-assembly like you think it is, they are explaining the various ways of folding these peptide chains into the tertiary structured proteins. They mention pathogenic folding, or in other words, undesirably folded proteins. This occurs when there is not proper post-translational folding. As per the example used in the paper you provided, amyloid fibrils are the pathogenic correlate of Alzheimer's disease. Another example they use is viral capsid formation. But this isn't necessarily self-assembly as you think, it still requires the host cell's machinery to forego transcription and translation to make the polypeptide sequence.
I do not doubt that tertiary and quaternary protein structures can self-assemble, in fact, biological organisms need chaperone proteins to literally chaperone the folding of the secondary protein structure (the polypeptide chain) into a coherent tertiary and quaternary structure. Failure to do so leads to all sorts of pathogenesis, i.e. amyloid fibrils.

So your wish that complex functional proteins can just self-assemble from scratch peptide monomers is totally unfounded in the literature. Transcription and translation are still required, and they can only parse sections of DNA that code for particular proteins... so the dilemma still remains, how could all of the proteins required for the muscle unit have been created by synchronous mutations? Especially considering the titin protein has over 100,000 base pairs (DNA units) of length to be coded for.
It is absolutely unfounded in the literature how such a synchronized event could have occurred to overcome the leap of irreducible complexity.
edit on 26-7-2019 by cooperton because: (no reason given)
new topics
-
President BIDEN's FBI Raided Donald Trump's Florida Home for OBAMA-NORTH KOREA Documents.
Political Conspiracies: 1 hours ago -
Maestro Benedetto
Literature: 2 hours ago -
Is AI Better Than the Hollywood Elite?
Movies: 3 hours ago -
Las Vegas UFO Spotting Teen Traumatized by Demon Creature in Backyard
Aliens and UFOs: 6 hours ago -
2024 Pigeon Forge Rod Run - On the Strip (Video made for you)
Automotive Discussion: 7 hours ago -
Gaza Terrorists Attack US Humanitarian Pier During Construction
Middle East Issues: 7 hours ago -
The functionality of boldening and italics is clunky and no post char limit warning?
ATS Freshman's Forum: 8 hours ago -
Meadows, Giuliani Among 11 Indicted in Arizona in Latest 2020 Election Subversion Case
Mainstream News: 9 hours ago -
Massachusetts Drag Queen Leads Young Kids in Free Palestine Chant
Social Issues and Civil Unrest: 9 hours ago -
Weinstein's conviction overturned
Mainstream News: 11 hours ago
top topics
-
President BIDEN's FBI Raided Donald Trump's Florida Home for OBAMA-NORTH KOREA Documents.
Political Conspiracies: 1 hours ago, 12 flags -
Krystalnacht on today's most elite Universities?
Social Issues and Civil Unrest: 12 hours ago, 9 flags -
University of Texas Instantly Shuts Down Anti Israel Protests
Education and Media: 15 hours ago, 8 flags -
Supreme Court Oral Arguments 4.25.2024 - Are PRESIDENTS IMMUNE From Later Being Prosecuted.
Above Politics: 12 hours ago, 8 flags -
Gaza Terrorists Attack US Humanitarian Pier During Construction
Middle East Issues: 7 hours ago, 7 flags -
Weinstein's conviction overturned
Mainstream News: 11 hours ago, 7 flags -
Massachusetts Drag Queen Leads Young Kids in Free Palestine Chant
Social Issues and Civil Unrest: 9 hours ago, 7 flags -
Meadows, Giuliani Among 11 Indicted in Arizona in Latest 2020 Election Subversion Case
Mainstream News: 9 hours ago, 5 flags -
Las Vegas UFO Spotting Teen Traumatized by Demon Creature in Backyard
Aliens and UFOs: 6 hours ago, 4 flags -
2024 Pigeon Forge Rod Run - On the Strip (Video made for you)
Automotive Discussion: 7 hours ago, 3 flags
active topics
-
One Flame Throwing Robot Dog for Christmas Please!
Weaponry • 11 • : Therealbeverage -
British TV Presenter Refuses To Use Guest's Preferred Pronouns
Education and Media • 167 • : WakeUpBeer -
University of Texas Instantly Shuts Down Anti Israel Protests
Education and Media • 257 • : DBCowboy -
HORRIBLE !! Russian Soldier Drinking Own Urine To Survive In Battle
World War Three • 48 • : Therealbeverage -
The Good News According to Jesus - Episode 1
Religion, Faith, And Theology • 3 • : Therealbeverage -
Weinstein's conviction overturned
Mainstream News • 23 • : Therealbeverage -
Supreme Court Oral Arguments 4.25.2024 - Are PRESIDENTS IMMUNE From Later Being Prosecuted.
Above Politics • 82 • : Lumenari -
President BIDEN's FBI Raided Donald Trump's Florida Home for OBAMA-NORTH KOREA Documents.
Political Conspiracies • 2 • : Boomer1947 -
Is AI Better Than the Hollywood Elite?
Movies • 6 • : Therealbeverage -
Mood Music Part VI
Music • 3103 • : TheDiscoKing