It looks like you're using an Ad Blocker.
Please white-list or disable AboveTopSecret.com in your ad-blocking tool.
Thank you.
Some features of ATS will be disabled while you continue to use an ad-blocker.
12
share:
Wikipedia
Molecular phylogenetics /məˈlɛkjʊlər faɪlɵdʒɪˈnɛtɪks/ is the analysis of hereditary molecular differences, mainly in DNA sequences, to gain information on an organism's evolutionary relationships. The result of a molecular phylogenetic analysis is expressed in a phylogenetic tree. Molecular phylogenetics is one aspect of molecular systematics, a broader term that also includes the use of molecular data in taxonomy and biogeography.
I was a little bit bored today, so I decided to conduct a small phylogenetic study. The species I chose to study included human, chimpanzee, gorilla, and orangutang. I'll describe briefly what I did (very standard practices), and attempt to explain the results. I'm going to leave out a lot of specifics, but feel free to ask.
First, I acquired the genomes of the aforementioned species. More specifically, I did not download the entire nucleotide sequences of all the chromosomes, but instead the amino acid sequences of all the proteins that have been detected/predicted from these genomes and curated manually by humans. Such files are publicly available in for example here, e.g. protein.fa.gz file in this directory includes all the predicted proteins of the orangutang genome.
After extracting the files, I build a database from them by utilizing the NCBI BLAST+ program. Subsequently, in order to identify which protein sequences in all the genomes represented the same genes, I used the same BLAST program (BLASTP search). This resulted in a file that was about 3.5 GB in size, and had scores for 'all vs. all' searches. Next, to parse this file, and to identify the clusters of similar genes, I utilized the orthomcl pipeline. After this, I had a file in which all the orthologs, co-orthologs, and inparalogs were grouped (see this for more info).
Now, many groups included numerous genes from the same organisms, which is to be expected because of gene duplication, which is one of the main mechanisms of how new functional genes are acquired. Below is an example of the third largest group (it looks like these genes multiplied a lot especially in the chimp lineage):
Gor|ENSGGOP00000018953 Gor|ENSGGOP00000019699 Gor|ENSGGOP00000023909 Gor|ENSGGOP00000026856 Gor|ENSGGOP00000009558 Gor|ENSGGOP00000018054 Gor|ENSGGOP00000006228 Gor|ENSGGOP00000014501 Gor|ENSGGOP00000000105 Gor|ENSGGOP00000000136 Gor|ENSGGOP00000004428 Gor|ENSGGOP00000004073 Gor|ENSGGOP00000024079 Gor|ENSGGOP00000026350 Gor|ENSGGOP00000005189 Gor|ENSGGOP00000021742 Gor|ENSGGOP00000001566 Pan|XP_001145156.1 Pan|XP_003316313.1 Pan|XP_524215.2 Pan|XP_003339367.1 Pan|XP_003316766.1 Pan|XP_003316765.1 Pan|XP_003316311.1 Pan|XP_003316775.1 Pan|XP_003316777.1 Pan|XP_003316779.1 Pan|XP_003316778.1 Pan|XP_003316776.1 Pan|XP_003316821.1 Pan|XP_512932.2 Pan|XP_524427.2 Pan|XP_001143794.1 Pan|XP_001145312.1 Pan|XP_003316749.1 Pan|XP_003316750.1 Pan|XP_003316793.1 Pan|XP_003316794.1 Pan|XP_003316780.1 Pan|XP_003316781.1 Pan|XP_003316770.1 Pan|XP_512935.3 Pan|XP_003316773.1 Pan|XP_003316771.1 Pan|XP_003316772.1 Pan|XP_003316774.1 Pan|XP_003316768.1 Pan|XP_003316769.1 Pan|XP_003316767.1 Pan|XP_003316782.1 Pan|XP_003316783.1 Pon|XP_003779321.1 Pon|XP_002829095.2 Pon|XP_003779185.1 Pon|XP_002829906.1 Pon|XP_003779320.1 Pon|XP_002829923.2 Pon|NP_001125458.1 Pon|XP_002829915.1 Pon|XP_002829919.1 Pon|XP_002829916.1 Pon|XP_002829920.1 Pon|XP_003779323.1 Pon|XP_003779322.1 Pon|XP_002829921.1 Hom|NP_612356.1 Hom|NP_065708.2 Hom|NP_065931.3 Hom|NP_694995.2 Hom|NP_001166244.1 Hom|NP_690873.2 Hom|NP_001018855.2 Hom|NP_001186224.1 Hom|NP_005764.2 Hom|NP_060349.1 Hom|NP_942152.1 Hom|NP_001010879.2 Hom|NP_689688.2 Hom|NP_116217.1 Hom|NP_001252526.1 Hom|NP_001252527.1 Hom|NP_006376.2 Hom|NP_001252528.1 Hom|NP_001252529.1 Hom|NP_787068.3 Hom|NP_001191747.1 Hom|NP_079038.2 Hom|NP_775903.3 Hom|NP_001191746.1 Gor|ENSGGOP00000003307 Gor|ENSGGOP00000016362 Pon|XP_003776342.1 Gor|ENSGGOP00000014292 Pon|XP_003779325.1 Gor|ENSGGOP00000025158 Pan|XP_001142575.2 Gor|ENSGGOP00000017703 Gor|ENSGGOP00000025564 Gor|ENSGGOP00000002632 Pon|XP_003779772.1 Pan|XP_003316788.1 Pon|XP_003779326.1
To see what these genes are, you can go here and paste the identifiers which begin with XP/NP into the search field (this doesn't work for the gorilla sequences (Gor) because I acquired them from elsewhere and they're not reference sequences).
Now, I selected randomly groups which only had one gene from each organism. From these sequences, I attempted to resolve phylogeny. In this step, I 1) aligned the sequences with muscle, 2) removed gap regions from the alignments with Gblocks, 3) sought the best amino acid substitution models with prottest, 4) constructed bootstrapped maximum-likelihood trees with RaxML, and 5) visualized the trees with FigTree.
Below is the first group I randomly selected (I used rat as an outgroup, the sequence I got by doing a BLASTP of the human sequence against the NCBI database):
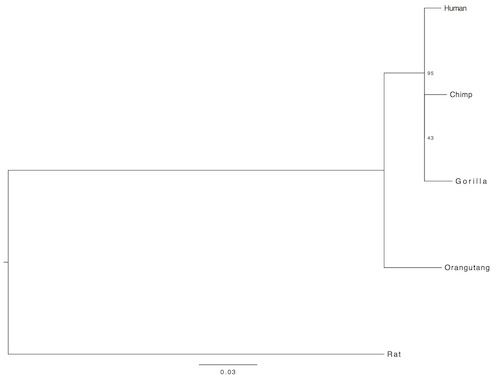
For the second randomly selected group, I used dog as the outgroup:
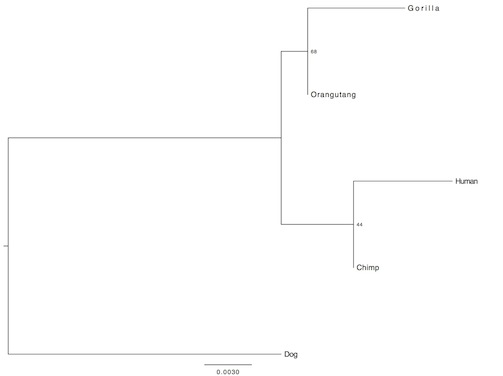
In the third case, my outgroup was horse:
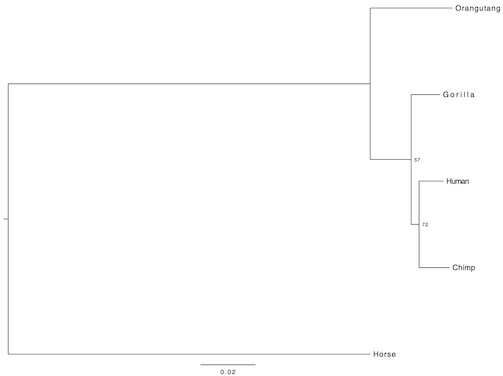
Now as you see, the evolutionary relation of the included species appears to be a little bit different in all the pictures (the third is the most similar to accepted phylogeny). Additionally, the bootsrap values are rather low. Why? Because single gene analyses offer in general very low resolution. There is no mechanism that drives the same gene to change at the same pace in different organisms. So, for my final analysis, in order to increase resolution, I concatenated further 10 randomly selected genes. This time, I did not include an outgroup, instead, I rooted the tree from the midpoint:
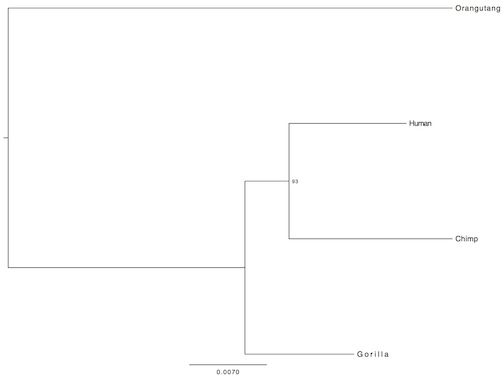
The result was to be expected. Below is the same tree, except in different style:
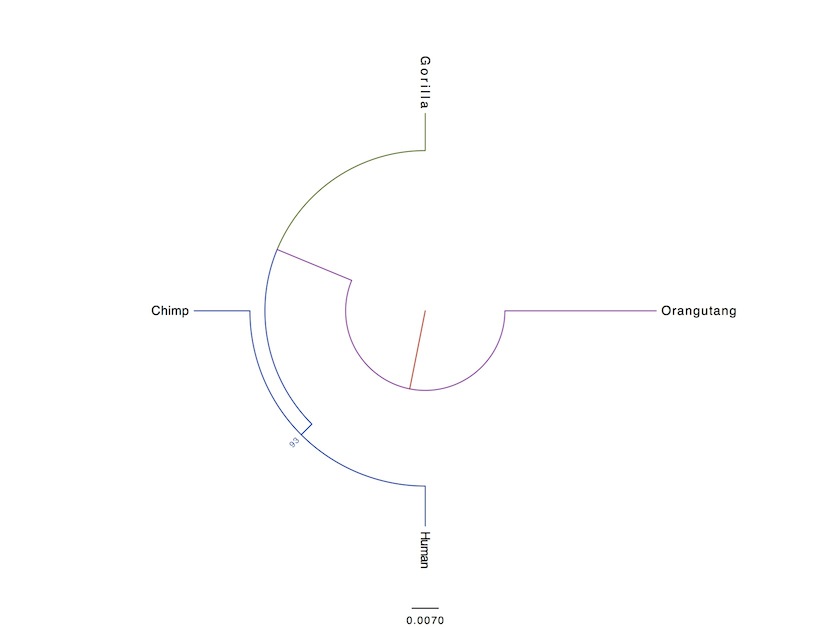
The red in the center represents the common ancestor of chimps, gorillas, humans, and orangutangs. So first, orangutang lineage separates from the chimp/human/gorilla lineage. Next, where purple, green and blue meet, the gorilla lineage separates from the human/chimp lineage, then finally, about 6 million years ago (number is not from my analysis), the human and chimp lineages separated. Also, take notice that on the basis of these 10 genes, humans are genetically more similar to orangutangs than chimps are.
This is what the data says. Ape pride world wide
edit on 19-10-2012 by rhinoceros because: (no reason given)
reply to post by rhinoceros
Very interesting, and great job taking the time to do this! Even if the fossil record is incomplete, molecular clocks can be used to reconstruct phylogenies via genetic distances which are based on constant rates.
But the question is, should humans be classified in the same family as the great apes, or should they belong to their own family?
I would rather have more in common with primates, than be related to two individuals who fell from a fixed state of grace into sin.
Very interesting, and great job taking the time to do this! Even if the fossil record is incomplete, molecular clocks can be used to reconstruct phylogenies via genetic distances which are based on constant rates.
But the question is, should humans be classified in the same family as the great apes, or should they belong to their own family?
I would rather have more in common with primates, than be related to two individuals who fell from a fixed state of grace into sin.
Originally posted by IEtherianSoul9
But the question is, should humans be classified in the same family as the great apes, or should they belong to their own family?
Humans are classified in the same family as chimps, bonobos, gorillas, and orangutangs, i.e. Hominidae. Humans are also classified in the same subfamily as chimps, bonobos and gorillas, i.e. Homininae. Further, humans are classified in the same tribe as chimps and bonobos, i.e. Hominini. However, in the end these distinctions are subjective. For example, if we applied the same species criteria that we apply to bacteria to animals, everything between humans and lemurs would belong to the same species..
I cant even begin to understand what you just posted. But I like the pictures lol.
Genetics and computer programing are 2 things I wish I had the capacity to learn as I think they are one and the same thing. Gene's are just chemical code for organic machines just as much as blocks of electromagnetic code are for digital machines.
Awesome work none the less. Keep it up and keep experimenting. 6 million year split from chimps huh? Are you sure we havent devolved backwards? lol
Genetics and computer programing are 2 things I wish I had the capacity to learn as I think they are one and the same thing. Gene's are just chemical code for organic machines just as much as blocks of electromagnetic code are for digital machines.
Awesome work none the less. Keep it up and keep experimenting. 6 million year split from chimps huh? Are you sure we havent devolved backwards? lol
Originally posted by TiM3LoRd
Genetics and computer programing are 2 things I wish I had the capacity to learn as I think they are one and the same thing. Gene's are just chemical code for organic machines just as much as blocks of electromagnetic code are for digital machines.
Back when I studied computer science, I used to think like this. However, it's not really true. The workings of the cell cannot be understood by simply studying its DNA and the genes it holds. There is no "source code" for a program in DNA. More like, the program is a massively complex network of interactions between DNA, RNA, proteins and other organic and inorganic molecules. Understanding these networks and their working is one of the greatest challenges in biology..
Originally posted by rhinoceros
Originally posted by TiM3LoRd
Genetics and computer programing are 2 things I wish I had the capacity to learn as I think they are one and the same thing. Gene's are just chemical code for organic machines just as much as blocks of electromagnetic code are for digital machines.
Back when I studied computer science, I used to think like this. However, it's not really true. The workings of the cell cannot be understood by simply studying its DNA and the genes it holds. There is no "source code" for a program in DNA. More like, the program is a massively complex network of interactions between DNA, RNA, proteins and other organic and inorganic molecules. Understanding these networks and their working is one of the greatest challenges in biology..
yes well obviously the organic machine has had billions of years to evolve and grow in complexity. given the same time frame I think the digital machines will reach the same level of complexity maybe more.
Originally posted by rhinoceros
Originally posted by TiM3LoRd
Genetics and computer programing are 2 things I wish I had the capacity to learn as I think they are one and the same thing. Gene's are just chemical code for organic machines just as much as blocks of electromagnetic code are for digital machines.
Back when I studied computer science, I used to think like this. However, it's not really true. The workings of the cell cannot be understood by simply studying its DNA and the genes it holds. There is no "source code" for a program in DNA. More like, the program is a massively complex network of interactions between DNA, RNA, proteins and other organic and inorganic molecules. Understanding these networks and their working is one of the greatest challenges in biology..
And enter the physicists. Quantum biology has opened doors to biologists and they're now in a position to answer questions that were unthinkable not all that long ago. I really wish I'd paid more attention to physics in school.
new topics
-
4/27/24 New Jersey Earthquake
Fragile Earth: 4 hours ago -
Fun with extreme paints
Interesting Websites: 5 hours ago -
CIA is alleged to be operat social media troll frms in Kyiv
ATS Skunk Works: 7 hours ago -
Rainbow : Stargazer
Music: 7 hours ago -
I sleep no more.
Philosophy and Metaphysics: 10 hours ago -
Canada caught red-handed manipulating live weather data and make it warmer
Fragile Earth: 10 hours ago -
Why Files Our Alien Overlords | How We Secretly Serve The Tall Whites
Aliens and UFOs: 11 hours ago
top topics
-
Canada caught red-handed manipulating live weather data and make it warmer
Fragile Earth: 10 hours ago, 16 flags -
Why Files Our Alien Overlords | How We Secretly Serve The Tall Whites
Aliens and UFOs: 11 hours ago, 11 flags -
Curse of King Tuts Tomb Solved
Ancient & Lost Civilizations: 13 hours ago, 9 flags -
CIA is alleged to be operat social media troll frms in Kyiv
ATS Skunk Works: 7 hours ago, 6 flags -
4/27/24 New Jersey Earthquake
Fragile Earth: 4 hours ago, 6 flags -
What allies does Trump have in the world?
ATS Skunk Works: 13 hours ago, 5 flags -
I sleep no more.
Philosophy and Metaphysics: 10 hours ago, 4 flags -
Fun with extreme paints
Interesting Websites: 5 hours ago, 2 flags -
Rainbow : Stargazer
Music: 7 hours ago, 1 flags
active topics
-
Ireland VS Globalists
Social Issues and Civil Unrest • 14 • : covent -
Gov Kristi Noem Shot and Killed "Less Than Worthless Dog" and a 'Smelly Goat
2024 Elections • 92 • : cherokeetroy -
Today I am 8
Members • 21 • : randomuser2034 -
Big Storms
Fragile Earth • 19 • : lilzazz -
George Knapp AMA on DI
Area 51 and other Facilities • 40 • : TheDiscoKing -
I sleep no more.
Philosophy and Metaphysics • 13 • : RAY1990 -
Silent Moments --In Memory of Beloved Member TDDA
Short Stories • 76 • : RAY1990 -
Why Files Our Alien Overlords | How We Secretly Serve The Tall Whites
Aliens and UFOs • 11 • : TheValeyard -
What allies does Trump have in the world?
ATS Skunk Works • 6 • : nugget1 -
CIA is alleged to be operat social media troll frms in Kyiv
ATS Skunk Works • 23 • : theatreboy
12
