It looks like you're using an Ad Blocker.
Please white-list or disable AboveTopSecret.com in your ad-blocking tool.
Thank you.
Some features of ATS will be disabled while you continue to use an ad-blocker.
share:
a reply to: cooperton
Let me remind you that YOU never proved your interpretation. You merely post a jpg, rework it and expect everyone to accept your inept way of describing the system. You don't even understand the electron transport system. Your description is 100% wrong. Go back to school (I recommend starting in the 1st grade) and learn science properly.
Regardless of that, you still need to either prove that somehow the electron transport chain can work without ATP synthase, or admit that it is irreducibly complex.
Let me remind you that YOU never proved your interpretation. You merely post a jpg, rework it and expect everyone to accept your inept way of describing the system. You don't even understand the electron transport system. Your description is 100% wrong. Go back to school (I recommend starting in the 1st grade) and learn science properly.
originally posted by: cooperton
originally posted by: Phantom423
Please cite a textbook, research article or other professional material which says that evolution is about one organism changing into another. And give an example like: A mouse turning into an elephant or a bedbug turning into a dog.
Thank you.
Evolutionary theory insists that organisms have been evolving into other organisms over time. We never observe this. Populations of organisms cannot change into another over time. Your faith that they do is unfounded in empirical science
Cooperton, I never thought I'd see the day when you admit to being a fraud. I guess the Silence of the Lambs says it all.

Your pal's jail cell is empty at the moment. Perhaps you'd like to start your sentence early for potential early release?
en.wikipedia.org...
edit on 7-6-2020 by Phantom423 because: (no reason given)
originally posted by: Phantom423
Please cite a textbook, research article or other professional material which says that evolution is about one organism changing into another.
Ape-like creatures were theorized to have changed into another organism over time - home sapiens. The entire point of evolutionary theory is that all organisms mutated from the progenitor first living organism.
Let me remind you that YOU never proved your interpretation
Yikes... it was such basic biology I would've figured you knew it. The electron transport chain needs ATP synthase to generate ATP from the electrochemical gradient that emerged from the preceding metabolic processes. Since all the complexes of the electron transport chain need each other to have a fully functioning metabolism, it can not work properly without ATP synthase. Therefore, the electron transport chain is irreducibly complex
edit on 7-6-2020 by cooperton because: (no reason given)
originally posted by: cooperton
originally posted by: Phantom423
Please cite a textbook, research article or other professional material which says that evolution is about one organism changing into another.
Ape-like creatures were theorized to have changed into another organism over time - home sapiens. The entire point of evolutionary theory is that all organisms mutated from the progenitor first living organism.
Let me remind you that YOU never proved your interpretation
Yikes... it was such basic biology I would've figured you knew it. The electron transport chain needs ATP synthase to generate ATP from the electrochemical gradient that emerged from the preceding metabolic processes. Since all the complexes of the electron transport chain need each other to have a fully functioning metabolism, it can not work properly without ATP synthase. Therefore, the electron transport chain is irreducibly complex
You avoid answering the question like the plague. The reason is because there is no textbook, professional journal or research article that ever said that an ape turned into a man. You think you can fool everyone by inserting terms like "over time", etc. Well you don't. You fool no one.
And no, you don't understand the physics of the electron transport system. You're making it up as you go along inserting crap along the way. If you understood anything about the electron transport system you would know that it is an excellent example of evolution itself.
edit on 7-6-2020 by Phantom423 because: (no reason given)
originally posted by: Phantom423
there is no textbook, professional journal or research article that ever said that an ape turned into a man.
You know thats not what I said. When I refer to an "ape-like creature" that is a reference to the theorized hominids that preceded the evolution of homo sapiens.
You think you can fool everyone by inserting terms like "over time", etc. Well you don't. You fool no one.
You need to relax
And no, you don't understand the physics of the electron transport system.
Where was I incorrect in my analysis? Do you have a solution to how it could work without ATP synthase? The electron transport chain needs ATP synthase to function, among many other required proteins, therefore it is irreducibly complex
I'll take the inability to answer the question as a total FAIL. Question remains unanswered.
You're a weasel who squirms around the obvious in an attempt to obscure the truth. That's a definition of a fraud.
As for the electron transport system, you have zero understanding of its development, its components or why it's even there.
During evolution, proton pumps have arisen independently on multiple occasions. Thus, not only throughout nature but also within single cells, different proton pumps that are evolutionarily unrelated can be found. Proton pumps are divided into different major classes of pumps that use different sources of energy, have different polypeptide compositions and evolutionary origins.
What this means is that all proton pumps are component systems. They evolved from primitive proton pumps to more complex structures when more complexity to support the organism was required.
There is no "irreducible complexity" here. It's a series of systems built over time to accommodate life. ATP synthase itself has several molecular structures. And an enzyme complex may not always be required to produce ATP.
Your explanation is based on Michael Behe's work which was proven in court to be unsubstantiated. An irreducible complex system would have no precursors and would be a stand-alone, unique structure with no precedence. The electron transport system as we know it today does not fill that definition. What's more, it will continue to evolve to accommodate whatever the lifeform requires.
You failed again, Cooperton. I would hate to be you presenting my report card to Mom and Dad.
I will have to enlarge these jpgs to make them readable
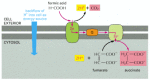
The oxidation of formic acid in some present-day bacteria
In such anaerobic bacteria, including E. coli, the oxidation is mediated by an energy-conserving electron-transport chain in the plasma membrane. As indicated, the starting materials are formic acid and fumarate, and the products are succinate and CO2. Note that H+ is consumed inside the cell and generated outside the cell, which is equivalent to pumping H+ to the cell exterior. Thus, this membrane-bound electron-transport system can generate an electrochemical proton gradient across the plasma membrane. The redox potential of the formic acid-CO2 pair is -420 mV, while that of the fumarate-succinate pair is +30 mV.


You're a weasel who squirms around the obvious in an attempt to obscure the truth. That's a definition of a fraud.
As for the electron transport system, you have zero understanding of its development, its components or why it's even there.
During evolution, proton pumps have arisen independently on multiple occasions. Thus, not only throughout nature but also within single cells, different proton pumps that are evolutionarily unrelated can be found. Proton pumps are divided into different major classes of pumps that use different sources of energy, have different polypeptide compositions and evolutionary origins.
Early cells are believed to have been bacteriumlike organisms living in an environment rich in highly reduced organic molecules that had been formed by geochemical processes over the course of hundreds of millions of years. They may have derived most of their ATP by converting these reduced organic molecules to a variety of organic acids, which were then released as waste products. By acidifying the environment, these fermentations may have led to the evolution of the first membrane-bound H+ pumps, which could maintain a neutral pH in the cell interior by pumping out H+.
The properties of present-day bacteria suggest that an electron-transport-driven H+ pump and an ATP-driven H+ pump first arose in this anaerobic environment. Reversal of the ATP-driven pump would have allowed it to function as an ATP synthase.As more effective electron-transport chains developed, the energy released by redox reactions between inorganic molecules and/or accumulated nonfermentable compounds produced a large electrochemical proton gradient, which could be harnessed by the ATP-driven pump for ATP production.
Because preformed organic molecules were replenished only very slowly by geochemical processes, the proliferation of bacteria that used them as the source of both carbon and reducing power could not go on forever. The depletion of fermentable organic nutrients presumably led to the evolution of bacteria that could use CO2to make carbohydrates. By combining parts of the electron-transport chains that had developed earlier, light energy was harvested by a single photosystem in photosynthetic bacteria to generate the NADPH required for carbon fixation. The subsequent appearance of the more complex photosynthetic electron-transport chains of the cyanobacteria allowed H2O to be used as the electron donor for NADPH formation, rather than the much less abundant electron donors required by other photosynthetic bacteria. Life could then proliferate over large areas of the Earth, so that reduced organic molecules accumulated again.
About 2×109 years ago, the O2 released by photosynthesis in cyanobacteria began to accumulate in the atmosphere. Once both organic molecules and O2 had become abundant, electron-transport chains became adapted for the transport of electrons from NADH to O2, and efficient aerobic metabolism developed in many bacteria. Exactly the same aerobic mechanisms operate today in the mitochondria of eucaryotes, and there is increasing evidence that both mitochondria and chloroplasts evolved from aerobic bacteria that were endocytosed by primitive eucaryotic cells.
What this means is that all proton pumps are component systems. They evolved from primitive proton pumps to more complex structures when more complexity to support the organism was required.
There is no "irreducible complexity" here. It's a series of systems built over time to accommodate life. ATP synthase itself has several molecular structures. And an enzyme complex may not always be required to produce ATP.
Your explanation is based on Michael Behe's work which was proven in court to be unsubstantiated. An irreducible complex system would have no precursors and would be a stand-alone, unique structure with no precedence. The electron transport system as we know it today does not fill that definition. What's more, it will continue to evolve to accommodate whatever the lifeform requires.
You failed again, Cooperton. I would hate to be you presenting my report card to Mom and Dad.
I will have to enlarge these jpgs to make them readable

The oxidation of formic acid in some present-day bacteria
In such anaerobic bacteria, including E. coli, the oxidation is mediated by an energy-conserving electron-transport chain in the plasma membrane. As indicated, the starting materials are formic acid and fumarate, and the products are succinate and CO2. Note that H+ is consumed inside the cell and generated outside the cell, which is equivalent to pumping H+ to the cell exterior. Thus, this membrane-bound electron-transport system can generate an electrochemical proton gradient across the plasma membrane. The redox potential of the formic acid-CO2 pair is -420 mV, while that of the fumarate-succinate pair is +30 mV.





edit on 9-6-2020 by Phantom423 because: (no reason given)
edit on 9-6-2020 by Phantom423 because: (no
reason given)
The jpgs come out of a book so the illustrations can't be enlarged. The captions are posted beneath the illustration.
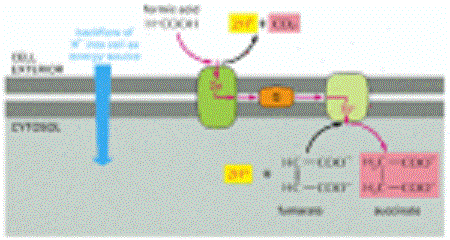
The oxidation of formic acid in some present-day bacteria
In such anaerobic bacteria, including E. coli, the oxidation is mediated by an energy-conserving electron-transport chain in the plasma membrane. As indicated, the starting materials are formic acid and fumarate, and the products are succinate and CO2. Note that H+ is consumed inside the cell and generated outside the cell, which is equivalent to pumping H+ to the cell exterior. Thus, this membrane-bound electron-transport system can generate an electrochemical proton gradient across the plasma membrane. The redox potential of the formic acid-CO2 pair is -420 mV, while that of the fumarate-succinate pair is +30 mV.
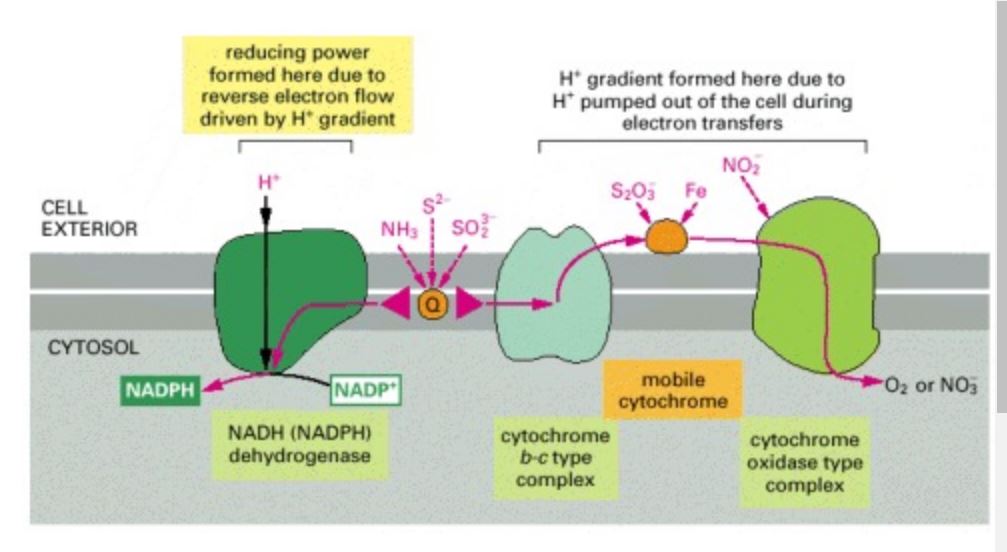
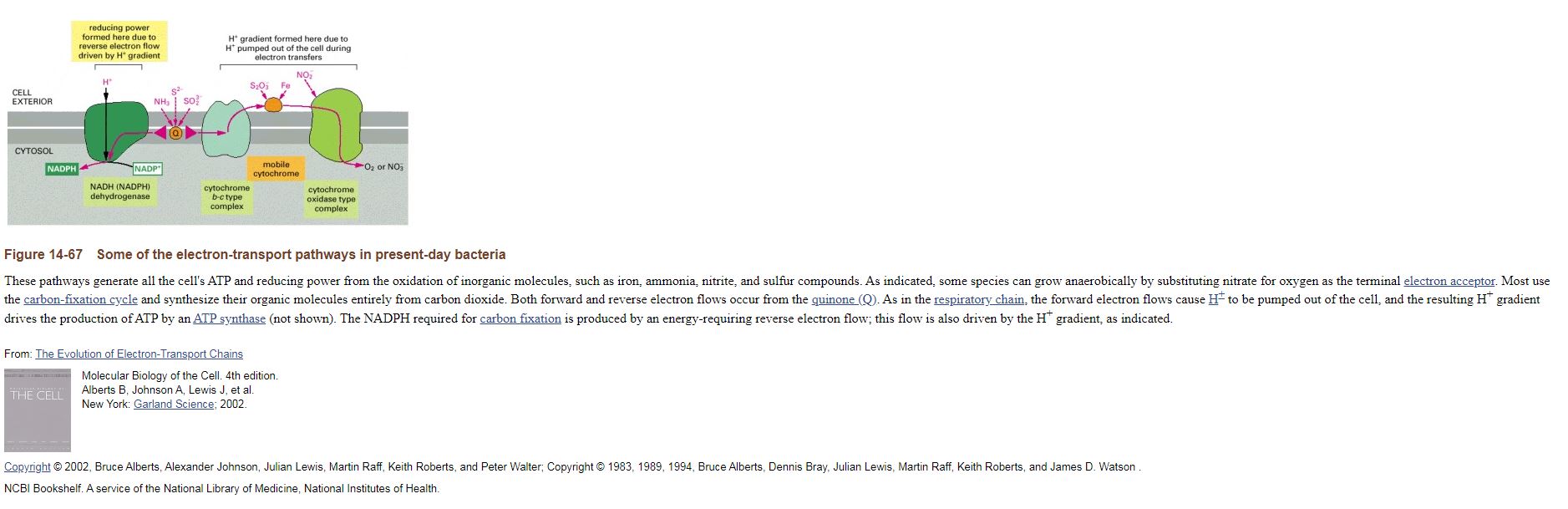
Some of the electron-transport pathways in present-day bacteria
These pathways generate all the cell's ATP and reducing power from the oxidation of inorganic molecules, such as iron, ammonia, nitrite, and sulfur compounds. As indicated, some species can grow anaerobically by substituting nitrate for oxygen as the terminal electron acceptor. Most use the carbon-fixation cycle and synthesize their organic molecules entirely from carbon dioxide. Both forward and reverse electron flows occur from the quinone (Q). As in the respiratory chain, the forward electron flows cause H+ to be pumped out of the cell, and the resulting H+ gradient drives the production of ATP by an ATP synthase (not shown). The NADPH required for carbon fixation is produced by an energy-requiring reverse electron flow; this flow is also driven by the H+ gradient, as indicated.
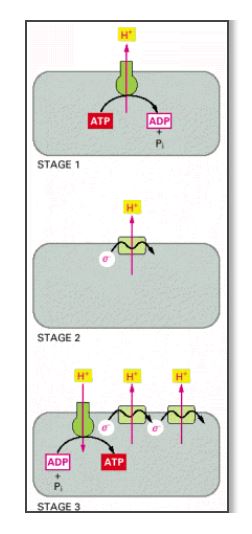
The evolution of oxidative phosphorylation mechanisms. One possible sequence is shown; the stages are described in the text.
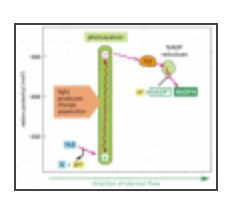
The general flow of electrons in a relatively primitive form of photosynthesis observed in present-day green sulfur bacteria
The photosystem in green sulfur bacteria resembles photosystem I in plants and cyanobacteria. Both photosystems use a series of iron-sulfur centers as the electron acceptors that eventually donate their high-energy electrons to ferredoxin (Fd). An example of a bacterium of this type is Chlorobium tepidum, which can thrive at high temperatures and low light intensities in hot springs.

Some major events that are believed to have occurred during the evolution of living organisms on Earth
With the evolution of the membrane-based process of photosynthesis, organisms could make their own organic molecules from CO2 gas. As explained in the text, the delay of more than 109 years between the appearance of bacteria that split water and released O2 during photosynthesis and the accumulation of high levels of O2 in the atmosphere is thought to be due to the initial reaction of the oxygen with abundant ferrous iron (Fe2+) dissolved in the early oceans. Only when the iron was used up would oxygen have started to accumulate in the atmosphere. In response to the rising levels of oxygen in the atmosphere, nonphotosynthetic oxygen-using organisms appeared, and the concentration of oxygen in the atmosphere leveled out.
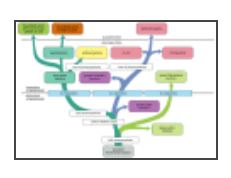
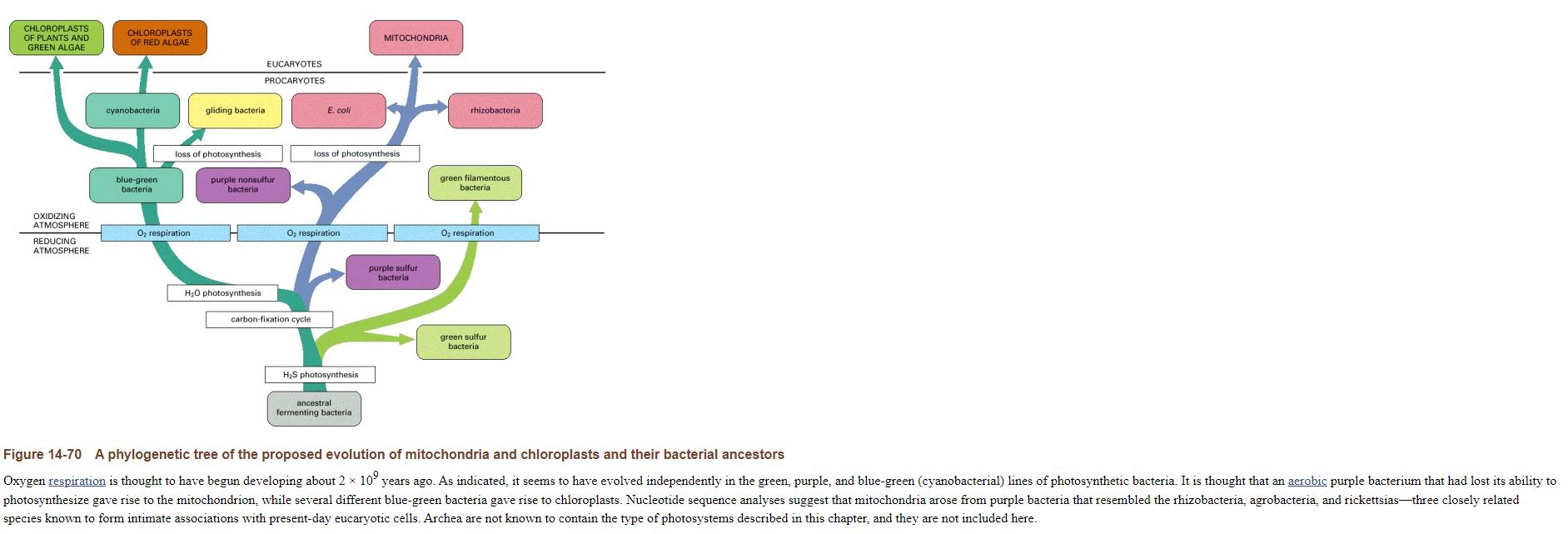
A phylogenetic tree of the proposed evolution of mitochondria and chloroplasts and their bacterial ancestors
Oxygen respiration is thought to have begun developing about 2 × 109 years ago. As indicated, it seems to have evolved independently in the green, purple, and blue-green (cyanobacterial) lines of photosynthetic bacteria. It is thought that an aerobic purple bacterium that had lost its ability to photosynthesize gave rise to the mitochondrion, while several different blue-green bacteria gave rise to chloroplasts. Nucleotide sequence analyses suggest that mitochondria arose from purple bacteria that resembled the rhizobacteria, agrobacteria, and rickettsias—three closely related species known to form intimate associations with present-day eucaryotic cells. Archea are not known to contain the type of photosystems described in this chapter, and they are not included here.
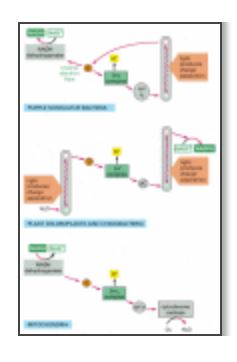
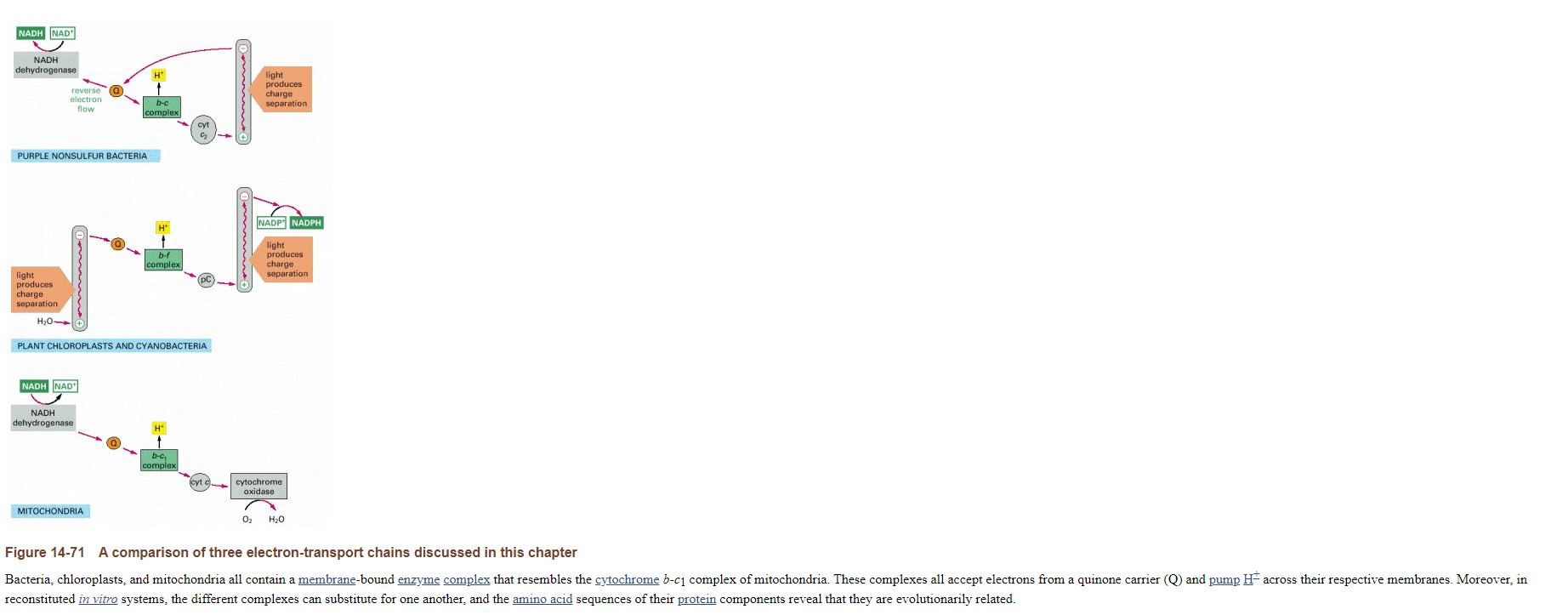
A comparison of three electron-transport chains discussed in this chapter
Bacteria, chloroplasts, and mitochondria all contain a membrane-bound enzyme complex that resembles the cytochrome b-c1 complex of mitochondria. These complexes all accept electrons from a quinone carrier (Q) and pump H+ across their respective membranes. Moreover, in reconstituted in vitro systems, the different complexes can substitute for one another, and the amino acid sequences of their protein components reveal that they are evolutionarily related.

The oxidation of formic acid in some present-day bacteria
In such anaerobic bacteria, including E. coli, the oxidation is mediated by an energy-conserving electron-transport chain in the plasma membrane. As indicated, the starting materials are formic acid and fumarate, and the products are succinate and CO2. Note that H+ is consumed inside the cell and generated outside the cell, which is equivalent to pumping H+ to the cell exterior. Thus, this membrane-bound electron-transport system can generate an electrochemical proton gradient across the plasma membrane. The redox potential of the formic acid-CO2 pair is -420 mV, while that of the fumarate-succinate pair is +30 mV.

Some of the electron-transport pathways in present-day bacteria
These pathways generate all the cell's ATP and reducing power from the oxidation of inorganic molecules, such as iron, ammonia, nitrite, and sulfur compounds. As indicated, some species can grow anaerobically by substituting nitrate for oxygen as the terminal electron acceptor. Most use the carbon-fixation cycle and synthesize their organic molecules entirely from carbon dioxide. Both forward and reverse electron flows occur from the quinone (Q). As in the respiratory chain, the forward electron flows cause H+ to be pumped out of the cell, and the resulting H+ gradient drives the production of ATP by an ATP synthase (not shown). The NADPH required for carbon fixation is produced by an energy-requiring reverse electron flow; this flow is also driven by the H+ gradient, as indicated.

The evolution of oxidative phosphorylation mechanisms. One possible sequence is shown; the stages are described in the text.

The general flow of electrons in a relatively primitive form of photosynthesis observed in present-day green sulfur bacteria
The photosystem in green sulfur bacteria resembles photosystem I in plants and cyanobacteria. Both photosystems use a series of iron-sulfur centers as the electron acceptors that eventually donate their high-energy electrons to ferredoxin (Fd). An example of a bacterium of this type is Chlorobium tepidum, which can thrive at high temperatures and low light intensities in hot springs.

Some major events that are believed to have occurred during the evolution of living organisms on Earth
With the evolution of the membrane-based process of photosynthesis, organisms could make their own organic molecules from CO2 gas. As explained in the text, the delay of more than 109 years between the appearance of bacteria that split water and released O2 during photosynthesis and the accumulation of high levels of O2 in the atmosphere is thought to be due to the initial reaction of the oxygen with abundant ferrous iron (Fe2+) dissolved in the early oceans. Only when the iron was used up would oxygen have started to accumulate in the atmosphere. In response to the rising levels of oxygen in the atmosphere, nonphotosynthetic oxygen-using organisms appeared, and the concentration of oxygen in the atmosphere leveled out.

A phylogenetic tree of the proposed evolution of mitochondria and chloroplasts and their bacterial ancestors
Oxygen respiration is thought to have begun developing about 2 × 109 years ago. As indicated, it seems to have evolved independently in the green, purple, and blue-green (cyanobacterial) lines of photosynthetic bacteria. It is thought that an aerobic purple bacterium that had lost its ability to photosynthesize gave rise to the mitochondrion, while several different blue-green bacteria gave rise to chloroplasts. Nucleotide sequence analyses suggest that mitochondria arose from purple bacteria that resembled the rhizobacteria, agrobacteria, and rickettsias—three closely related species known to form intimate associations with present-day eucaryotic cells. Archea are not known to contain the type of photosystems described in this chapter, and they are not included here.

A comparison of three electron-transport chains discussed in this chapter
Bacteria, chloroplasts, and mitochondria all contain a membrane-bound enzyme complex that resembles the cytochrome b-c1 complex of mitochondria. These complexes all accept electrons from a quinone carrier (Q) and pump H+ across their respective membranes. Moreover, in reconstituted in vitro systems, the different complexes can substitute for one another, and the amino acid sequences of their protein components reveal that they are evolutionarily related.
edit on 10-6-2020 by Phantom423 because: (no reason given)
edit on 10-6-2020 by Phantom423 because: (no reason given)
Why is nature so ordered? Why does science follow hard coded rules? Why is there not more chaos on this randomly evolved planet?
originally posted by: jjkenobi
Why is nature so ordered? Why does science follow hard coded rules? Why is there not more chaos on this randomly evolved planet?
In a word, survival. In biology it's called the structure-function relationship. The better the structure, the better the function. That's the idea of evolution. If systems do not evolve - all systems, not just ones that are "alive" - to meet the demands of their environment, they die off or disappear.
edit on 10-6-2020 by Phantom423 because: (no reason given)
a reply to: cooperton
Sometimes in life you get a second chance. Here's yours. Learn something.
www.abovetopsecret.com...
Sometimes in life you get a second chance. Here's yours. Learn something.
www.abovetopsecret.com...
originally posted by: Phantom423
During evolution, proton pumps have arisen independently on multiple occasions.
Show me empirical evidence that proton pumps can arise through random mutation. Your source admittedly makes faith-based assumptions - which is the opposite of science:
Early cells are believed to have been bacteriumlike organisms living in an environment rich in highly reduced organic molecules that had been formed by geochemical processes over the course of hundreds of millions of years. They may have derived most of their ATP by converting these reduced organic molecules to a variety of organic acids, which were then released as waste products. By acidifying the environment, these fermentations may have led to the evolution of the first membrane-bound H+ pumps, which could maintain a neutral pH in the cell interior by pumping out H+.
notice all the "may have's", and even their statement of faith: "believed to have been". It is faith, not science. They have the conclusion they want to prove, and then try to make stories to fit that narrative. It is backwards science.
Thus, not only throughout nature but also within single cells, different proton pumps that are evolutionarily unrelated can be found. Proton pumps are divided into different major classes of pumps that use different sources of energy, have different polypeptide compositions and evolutionary origins.
More faith. Never has it been seen in a lab that such a complex quaternary protein can come to be by the proposed evolutionary mechanisms. That wasn't even the debate either. The dilemma was that the electron transport chain needs all components to work.
The properties of present-day bacteria suggest that an electron-transport-driven H+ pump and an ATP-driven H+ pump first arose in this anaerobic environment. Reversal of the ATP-driven pump would have allowed it to function as an ATP synthase.
Yet this would be useless unless the rest of the electron transport chain were present, and if the rest of the electron transport chain were present, it would be hitherto useless because it didn't have ATP synthase. That is the paradox you can't wrap your head around because you're caught up in your biased dogma.
Once both organic molecules and O2 had become abundant, electron-transport chains became adapted for the transport of electrons from NADH to O2, and efficient aerobic metabolism developed in many bacteria. Exactly the same aerobic mechanisms operate today in the mitochondria of eucaryotes, and there is increasing evidence that both mitochondria and chloroplasts evolved from aerobic bacteria that were endocytosed by primitive eucaryotic cells.
cool story, but no empirical evidence to prove it. They are making creation myths. They take evolution as a presumed fact, and they make up a story to fit that narrative. Again, this is backwards science.
There is no "irreducible complexity" here. It's a series of systems built over time to accommodate life. ATP synthase itself has several molecular structures. And an enzyme complex may not always be required to produce ATP.
The oxidation of formic acid in some present-day bacteria
In such anaerobic bacteria, including E. coli, the oxidation is mediated by an energy-conserving electron-transport chain in the plasma membrane. As indicated, the starting materials are formic acid and fumarate, and the products are succinate and CO2. Note that H+ is consumed inside the cell and generated outside the cell, which is equivalent to pumping H+ to the cell exterior. Thus, this membrane-bound electron-transport system can generate an electrochemical proton gradient across the plasma membrane. The redox potential of the formic acid-CO2 pair is -420 mV, while that of the fumarate-succinate pair is +30 mV.
thanks for inadvertently proving my point. Even anaerobic microbes have a full electron transport chain - using sulfates, nitrates, or sulfur as the electron accepter rather than oxygen. This shows that even the most rudimentary organism requires a complete electron transport chain. No organism has ever been found that doesn't require ATP synthase, since this is true, this demonstrates that the electron transport chain can not function without ATP synthase - which was my original point. Your round-about answer resolved nothing except a lengthy specious speech.

"One possible sequence is shown".... Just more faith-based statements. No real science to prove what they're saying.
**I noticed with your edit you removed that snippet that said "one possible sequence is shown". Stop hiding.

Oh boy this is a good one... "some major events that are believe to have occurred
No empirical evidence, just faith.
It was a nice attempt, but all you posted was theoretical speculation without any proof of its validity. electron transport chains require ATP synthase, therefore making it irreducibly complex. You showed no example where an organism can live without ATP synthase. Instead you showed how much these people "believe" in these theorized mechanisms, yet cannot find actual proof for it. Therefore, it is not science.
edit on 10-6-2020 by
cooperton because: (no reason given)
a reply to: cooperton
Write the equation that says that proton pumps do not evolve. Empirical evidence is in the equations and the laboratory data. It's all there. You just refuse to acknowledge it. Everything posted above has laboratory evidence.
So go ahead. Give it your best shot. Do the math and the equations that prove your hypothesis.
Write the equation that says that proton pumps do not evolve. Empirical evidence is in the equations and the laboratory data. It's all there. You just refuse to acknowledge it. Everything posted above has laboratory evidence.
So go ahead. Give it your best shot. Do the math and the equations that prove your hypothesis.
edit on 10-6-2020 by Phantom423 because: (no reason given)
edit on 10-6-2020 by Phantom423 because: (no reason given)
a reply to: cooperton
P.S. I'm not posting ANYTHING until you prove unequivocally that your description is proven both mathematically and experimentally. As I remember, you're the one who says everything is impossible. So go ahead, make my day.
P.S. I'm not posting ANYTHING until you prove unequivocally that your description is proven both mathematically and experimentally. As I remember, you're the one who says everything is impossible. So go ahead, make my day.
edit on 10-6-2020 by Phantom423 because: (no reason given)
originally posted by: Phantom423
a reply to: cooperton
P.S. I'm not posting ANYTHING until you prove unequivocally that your description is proven both mathematically and experimentally. As I remember, you're the one who says everything is impossible. So go ahead, make my day.
I always know when I win an argument with you because you resort to demanding impractical things. Rather than discuss the straight-forward fact that all organisms require ATP synthase for the electron transport chain, you demand that I write a mathematical equation to prove a negative?? I'd love to.
ATP synthase is made of multiple sub-unit proteins. Each sub-unit protein is comprised of amino acid chains that form various functional groups that are meticulously necessary to generate proper form. The problem for evolution as that even generating one effective functional group via random mutation is estimated to be around 1/10,000,000,000,000,000,000,000,000,000,000,000...
000,000,000,000,000,000,000,000,000,000,000
according to this study: Source
To put things in perspective, the odds of winning the powerball lottery are 1/292,000,000
Now that super large number above is simply to arrive at some random functional group, let alone the exact functional group needed to create the desired function in that region of the amino acid sequence. Remember, each sub-unit requires many functional groups, and each ATP synthase requires many protein sub-units.
So if the probability for hitting a successful mutation to a functional group is approximately 1/10^67, and this would have to hit literally many, many times... therefore it is mathematically nearing absolute impossibility. About as possible as winning the lottery literally millions of times in a row. Not to mention that the rest of the electron transport chain is useless while it is desperately waiting for ATP synthase to randomly evolve... There is no way evolution can overcome these hurdles.
originally posted by: Phantom423
a reply to: cooperton
Answer the question. Do the math. Show the experimental data.
Can I play
Just fill me in so I can show you how stupid your argument is please Phantom
I love this
Pretending you understand science is your first clear and observable (I have tested it as well repetably) failure 🌈🦄💃
originally posted by: Phantom423
a reply to: cooperton
Answer the question. Do the math. Show the experimental data.
I just did. The odds of a functional group forming to create one component of a sub-unit of ATP synthase is approximately 1/10^67, and many of these functional groups are needed per sub-unit. That means the probability of ATP synthase evolving is nearly 0. You still haven't shown that the electron transport chain can work without ATP synthase, so therefore the electron transport chain is irreducibly complex.
originally posted by: jjkenobi
Why is nature so ordered? Why does science follow hard coded rules? Why is there not more chaos on this randomly evolved planet?
Exactly. A great question. Evolution is antithetical to the meticulously ordered universe we observe all around us. This shows that the theory simply does not belong. The physical laws are so precise, and so are biological organisms. We therefore must be the contrivance of a higher Intelligible Force
edit on 10-6-2020 by cooperton because: (no reason given)
a reply to: cooperton
Here's a list of references which contain the mathematical and experimental evidence. Pick one out and tell us why it's wrong. Then you can write a letter to the author.
References
General
Gesteland RF, Cech TR & Atkins JF (eds) (1999) The RNA World, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Hartwell L, Hood L, Goldberg ML et al. (2000) Genetics: from Genes to Genomes. Boston: McGraw Hill.
Lewin B (2000) Genes VII. Oxford: Oxford University Press.
Lodish H, Berk A, Zipursky SL et al. (2000) Molecular Cell Biology, 4th edn. New York: WH Freeman.
Stent GS (1971) Molecular Genetics: An Introductory Narrative. San Francisco: WH Freeman.
Stryer L (1995) Biochemistry, 4th edn. New York: WH Freeman.
Watson JD, Hopkins NH, Roberts JW et al. (1987) Molecular Biology of the Gene, 4th edn. Menlo Park, CA: Benjamin/Cummings.
From DNA to RNA
Berget SM , Moore C , Sharp PA . Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA. (1977);74:3171–3175. [PMC free article] [PubMed]
Black DL . Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. (2000);103:367–370. [PubMed]
Brenner S , Jacob F , Meselson M . An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. (1961);190:576–581. [PubMed]
Cech TR . Nobel lecture Self-splicing and enzymatic activity of an intervening sequence RNA from Tetrahymena. Biosci. Rep. (1990);10:239–261. [PubMed]
Chow LT , Gelinas RE , Broker TR . et al. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. (1977);12:1–8. [PubMed]
Conaway JW , Shilatifard A , Dvir A , Conaway RC . Control of elongation by RNA polymerase II. Trends Biochem. Sci. (2000);25:375–380. [PubMed]
Cramer P , Bushnell DA , Fu J . et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. (2000);288:640–649. [PubMed]
Crick F . Split genes and RNA splicing. Science. (1979);204:264–271. [PubMed]
Daneholt B . A look at messenger RNP moving through the nuclear pore. Cell. (1997);88:585–588. [PubMed]
Darnell JE Jr. RNA. Sci. Am. (1985);253(4):68–78. [PubMed]
Dvir A , Conaway JW , Conaway RC . Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. (2001);11:209–214. [PubMed]
Ebright RH . RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. (2000);304:687–698. [PubMed]
Eddy SR . Noncoding RNA genes. Curr. Opin. Genet. Dev. (1999);9:695–699. [PubMed]
Green MR . TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. (2000);25:59–63. [PubMed]
Harley CB , Reynolds RP . Analysis of E. coli promoter sequences. Nucleic Acids Res. (1987);15:2343–2361. [PMC free article] [PubMed]
Hirose Y , Manley JL . RNA polymerase II and the integration of nuclear events. Genes Dev. (2000);14:1415–1429. [PubMed]
Kadonaga JT . Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. (1998);92:307–313. [PubMed]
Lewis JD , Tollervey D . Like attracts like: getting RNA processing together in the nucleus. Science. (2000);288:1385–1389. [PubMed]
Lisser S , Margalit H . Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. (1993);21:1507–1516. [PMC free article] [PubMed]
Minvielle-Sebastia L , Keller W . mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. (1999);11:352–357. [PubMed]
Mooney RA , Landick R . RNA polymerase unveiled. Cell. (1999);98:687–690. [PubMed]
Olson MO , Dundr M , Szebeni A . The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. (2000);10:189–196. [PubMed]
Proudfoot N . Connecting transcription to messenger RNA processing. Trends Biochem. Sci. (2000);25:290–293. [PubMed]
Reed R . Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. (2000);12:340–345. [PubMed]
Roeder RG . The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. (1996);21:327–335. [PubMed]
Shatkin AJ , Manley JL . The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. (2000);7:838–842. [PubMed]
Smith CW , Valcarcel J . Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. (2000);25:381–388. [PubMed]
Staley JP , Guthrie C . Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. (1998);92:315–326. [PubMed]
Tarn WY , Steitz JA . Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. (1997);22:132–137. [PubMed]
von Hippel PH . An integrated model of the transcription complex in elongation, termination, and editing. Science. (1998);281:660–665. [PubMed]
From RNA to Protein
Abelson J , Trotta CR , Li H . tRNA splicing. J. Biol. Chem. (1998);273:12685–12688. [PubMed]
Anfinsen CB . Principles that govern the folding of protein chains. Science. (1973);181:223–230. [PubMed]
Cohen FE . Protein misfolding and prion diseases. J. Mol. Biol. (1999);293:313–320. [PubMed]
Crick FHC . The genetic code: III. Sci. Am. (1966);215(4):55–62. [PubMed]
Fedorov AN , Baldwin TO . Cotranslational protein folding. J. Biol. Chem. (1997);272:32715–32718. [PubMed]
Frank J . The ribosome—a macromolecular machine par excellence. Chem. Biol. (2000);7:R133–141. [PubMed]
Green R . Ribosomal translocation: EF-G turns the crank. Curr. Biol. (2000);10:R369–373. [PubMed]
Hartl FU . Molecular chaperones in cellular protein folding. Nature. (1996);381:571–580. [PubMed]
Hershko A , Ciechanover A , Varshavsky A . The ubiquitin system. Nat. Med. (2000);6:1073–1081. [PubMed]
Ibba M , Soll D . Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. (2000);69:617–650. [PubMed]
Kozak M . Initiation of translation in prokaryotes and eukaryotes. Gene. (1999);234:187–208. [PubMed]
Nissen P , Hansen J , Ban N . et al. The structural basis of ribosome activity in peptide bond synthesis. Science. (2000);289:920–930. [PubMed]
Nureki O , Vassylyev DG , Tateno M . et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. (1998);280:578–582. [PubMed]
Prusiner SB . Nobel lecture. Prions. Proc. Natl. Acad. Sci. USA. (1998);95:13363–13383. [PMC free article] [PubMed]
Rich A , Kim SH . The three-dimensional structure of transfer RNA. Sci. Am. (1978);238(1):52–62. [PubMed]
Sachs AB , Varani G . Eukaryotic translation initiation: there are (at least) two side to every story. Nat. Struct. Biol. (2000);7:356–361. [PubMed]
The Genetic Code. (1966) Cold Spring Harbor Symposium on Quantitative Biology, vol XXXI. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Turner GC , Varshavsky A . Detecting and measuring cotranslational protein degradation in vivo. Science. (2000);289:2117–2120. [PubMed]
Varshavsky A , Turner G , Du F . et al. The ubiquitin system and the N-end rule pathway. Biol. Chem. (2000);381:779–789. [PubMed]
Voges D , Zwickl P , Baumeister W . The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. (1999);68:1015–1068. [PubMed]
Here's a list of references which contain the mathematical and experimental evidence. Pick one out and tell us why it's wrong. Then you can write a letter to the author.
References
General
Gesteland RF, Cech TR & Atkins JF (eds) (1999) The RNA World, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Hartwell L, Hood L, Goldberg ML et al. (2000) Genetics: from Genes to Genomes. Boston: McGraw Hill.
Lewin B (2000) Genes VII. Oxford: Oxford University Press.
Lodish H, Berk A, Zipursky SL et al. (2000) Molecular Cell Biology, 4th edn. New York: WH Freeman.
Stent GS (1971) Molecular Genetics: An Introductory Narrative. San Francisco: WH Freeman.
Stryer L (1995) Biochemistry, 4th edn. New York: WH Freeman.
Watson JD, Hopkins NH, Roberts JW et al. (1987) Molecular Biology of the Gene, 4th edn. Menlo Park, CA: Benjamin/Cummings.
From DNA to RNA
Berget SM , Moore C , Sharp PA . Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA. (1977);74:3171–3175. [PMC free article] [PubMed]
Black DL . Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. (2000);103:367–370. [PubMed]
Brenner S , Jacob F , Meselson M . An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. (1961);190:576–581. [PubMed]
Cech TR . Nobel lecture Self-splicing and enzymatic activity of an intervening sequence RNA from Tetrahymena. Biosci. Rep. (1990);10:239–261. [PubMed]
Chow LT , Gelinas RE , Broker TR . et al. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. (1977);12:1–8. [PubMed]
Conaway JW , Shilatifard A , Dvir A , Conaway RC . Control of elongation by RNA polymerase II. Trends Biochem. Sci. (2000);25:375–380. [PubMed]
Cramer P , Bushnell DA , Fu J . et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. (2000);288:640–649. [PubMed]
Crick F . Split genes and RNA splicing. Science. (1979);204:264–271. [PubMed]
Daneholt B . A look at messenger RNP moving through the nuclear pore. Cell. (1997);88:585–588. [PubMed]
Darnell JE Jr. RNA. Sci. Am. (1985);253(4):68–78. [PubMed]
Dvir A , Conaway JW , Conaway RC . Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. (2001);11:209–214. [PubMed]
Ebright RH . RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. (2000);304:687–698. [PubMed]
Eddy SR . Noncoding RNA genes. Curr. Opin. Genet. Dev. (1999);9:695–699. [PubMed]
Green MR . TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. (2000);25:59–63. [PubMed]
Harley CB , Reynolds RP . Analysis of E. coli promoter sequences. Nucleic Acids Res. (1987);15:2343–2361. [PMC free article] [PubMed]
Hirose Y , Manley JL . RNA polymerase II and the integration of nuclear events. Genes Dev. (2000);14:1415–1429. [PubMed]
Kadonaga JT . Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. (1998);92:307–313. [PubMed]
Lewis JD , Tollervey D . Like attracts like: getting RNA processing together in the nucleus. Science. (2000);288:1385–1389. [PubMed]
Lisser S , Margalit H . Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. (1993);21:1507–1516. [PMC free article] [PubMed]
Minvielle-Sebastia L , Keller W . mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. (1999);11:352–357. [PubMed]
Mooney RA , Landick R . RNA polymerase unveiled. Cell. (1999);98:687–690. [PubMed]
Olson MO , Dundr M , Szebeni A . The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. (2000);10:189–196. [PubMed]
Proudfoot N . Connecting transcription to messenger RNA processing. Trends Biochem. Sci. (2000);25:290–293. [PubMed]
Reed R . Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. (2000);12:340–345. [PubMed]
Roeder RG . The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. (1996);21:327–335. [PubMed]
Shatkin AJ , Manley JL . The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. (2000);7:838–842. [PubMed]
Smith CW , Valcarcel J . Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. (2000);25:381–388. [PubMed]
Staley JP , Guthrie C . Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. (1998);92:315–326. [PubMed]
Tarn WY , Steitz JA . Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. (1997);22:132–137. [PubMed]
von Hippel PH . An integrated model of the transcription complex in elongation, termination, and editing. Science. (1998);281:660–665. [PubMed]
From RNA to Protein
Abelson J , Trotta CR , Li H . tRNA splicing. J. Biol. Chem. (1998);273:12685–12688. [PubMed]
Anfinsen CB . Principles that govern the folding of protein chains. Science. (1973);181:223–230. [PubMed]
Cohen FE . Protein misfolding and prion diseases. J. Mol. Biol. (1999);293:313–320. [PubMed]
Crick FHC . The genetic code: III. Sci. Am. (1966);215(4):55–62. [PubMed]
Fedorov AN , Baldwin TO . Cotranslational protein folding. J. Biol. Chem. (1997);272:32715–32718. [PubMed]
Frank J . The ribosome—a macromolecular machine par excellence. Chem. Biol. (2000);7:R133–141. [PubMed]
Green R . Ribosomal translocation: EF-G turns the crank. Curr. Biol. (2000);10:R369–373. [PubMed]
Hartl FU . Molecular chaperones in cellular protein folding. Nature. (1996);381:571–580. [PubMed]
Hershko A , Ciechanover A , Varshavsky A . The ubiquitin system. Nat. Med. (2000);6:1073–1081. [PubMed]
Ibba M , Soll D . Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. (2000);69:617–650. [PubMed]
Kozak M . Initiation of translation in prokaryotes and eukaryotes. Gene. (1999);234:187–208. [PubMed]
Nissen P , Hansen J , Ban N . et al. The structural basis of ribosome activity in peptide bond synthesis. Science. (2000);289:920–930. [PubMed]
Nureki O , Vassylyev DG , Tateno M . et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. (1998);280:578–582. [PubMed]
Prusiner SB . Nobel lecture. Prions. Proc. Natl. Acad. Sci. USA. (1998);95:13363–13383. [PMC free article] [PubMed]
Rich A , Kim SH . The three-dimensional structure of transfer RNA. Sci. Am. (1978);238(1):52–62. [PubMed]
Sachs AB , Varani G . Eukaryotic translation initiation: there are (at least) two side to every story. Nat. Struct. Biol. (2000);7:356–361. [PubMed]
The Genetic Code. (1966) Cold Spring Harbor Symposium on Quantitative Biology, vol XXXI. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Turner GC , Varshavsky A . Detecting and measuring cotranslational protein degradation in vivo. Science. (2000);289:2117–2120. [PubMed]
Varshavsky A , Turner G , Du F . et al. The ubiquitin system and the N-end rule pathway. Biol. Chem. (2000);381:779–789. [PubMed]
Voges D , Zwickl P , Baumeister W . The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. (1999);68:1015–1068. [PubMed]
new topics
-
What Am I Hearing
General Chit Chat: 28 minutes ago -
A Bunch of Maybe Drones Just Flew Across Hillsborough County
Aircraft Projects: 6 hours ago -
Who's coming with me?
General Conspiracies: 8 hours ago -
FBI Director CHRISTOPHER WRAY Will Resign Before President Trump Takes Office on 1.20.2025.
US Political Madness: 11 hours ago
top topics
-
The NIH is still sending taxpayer money to Chinese Labs to Conduct cruel animal experiments
Mainstream News: 12 hours ago, 12 flags -
FBI Director CHRISTOPHER WRAY Will Resign Before President Trump Takes Office on 1.20.2025.
US Political Madness: 11 hours ago, 9 flags -
A Bunch of Maybe Drones Just Flew Across Hillsborough County
Aircraft Projects: 6 hours ago, 7 flags -
Who's coming with me?
General Conspiracies: 8 hours ago, 4 flags -
Angela Merkel is terrified of dogs
Politicians & People: 13 hours ago, 3 flags -
What Am I Hearing
General Chit Chat: 28 minutes ago, 0 flags
active topics
-
-@TH3WH17ERABB17- -Q- ---TIME TO SHOW THE WORLD--- -Part- --44--
Dissecting Disinformation • 3626 • : duncanagain -
What Am I Hearing
General Chit Chat • 0 • : charlest2 -
Drones everywhere in New Jersey
Aliens and UFOs • 68 • : GENERAL EYES -
Rant. I am sick of people saying the police are revenue raising.
Rant • 9 • : inflaymes69 -
From the MUSK-RAMASWAMY Department of Government Efficiency commission - DOGE.
Above Politics • 61 • : WeMustCare -
A Bunch of Maybe Drones Just Flew Across Hillsborough County
Aircraft Projects • 15 • : Mantiss2021 -
Remember These Attacks When President Trump 2.0 Retribution-Justice Commences.
2024 Elections • 98 • : WeMustCare -
President-Elect DONALD TRUMP's 2nd-Term Administration Takes Shape.
Political Ideology • 297 • : WeMustCare -
Who's coming with me?
General Conspiracies • 27 • : AlroyFarms -
Will all hell break out? Jersey drones - blue beam
Aliens and UFOs • 39 • : Arbitrageur
