It looks like you're using an Ad Blocker.
Please white-list or disable AboveTopSecret.com in your ad-blocking tool.
Thank you.
Some features of ATS will be disabled while you continue to use an ad-blocker.
5
share:
SuperFrog posted this article for the library. I thought it was significant enough to the study of evolution that I created a special page for it at
the website. ats-library.wix.com...#!tree-of-life-project/c24p3
The project is open source - you can see the "tree" development here: tree.opentreeoflife.org...@1
I've made a request to be updated when new information or research is available.
Also, the ATS library has been extensively updated with new pages and a lot of videos. If anyone has any suggestions for additional articles or topics, please post in the sticky thread.
I am changing the GENOMICS topic to SPECIATION as I think that subject generates a lot of questions and includes genomics.
TREE OF LIFE PROJECT
Contributed by SuperFrog
"Tree of Life" for 2.3 Million Species Released
Large, open-access resource aims to be “Wikipedia” for evolutionary history
“Twenty five years ago people said this goal of huge trees was impossible,” Soltis said. “The Open Tree of Life is an important starting point that other investigators can now refine and improve for decades to come.”
September 18, 2015 | Robin A. Smith
Durham, NC - A first draft of the “tree of life” for the roughly 2.3 million named species of animals, plants, fungi and microbes -- from platypuses to puffballs -- has been released.
A collaborative effort among eleven institutions, the tree depicts the relationships among living things as they diverged from one another over time, tracing back to the beginning of life on Earth more than 3.5 billion years ago.
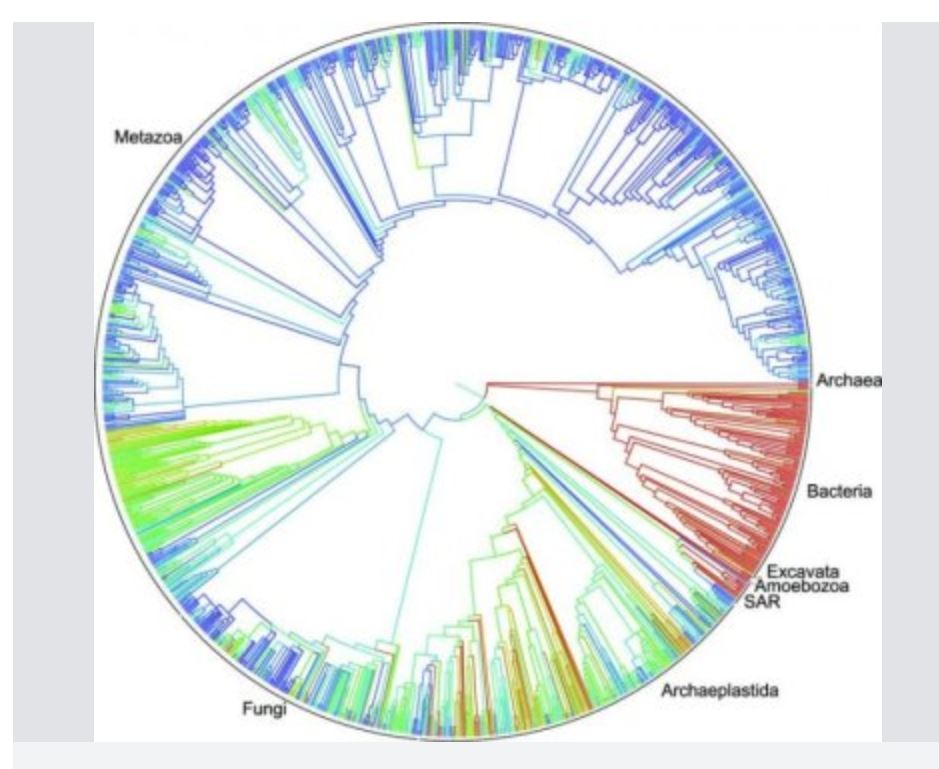
Tens of thousands of smaller trees have been published over the years for select branches of the tree of life -- some containing upwards of 100,000 species -- but this is the first time those results have been combined into a single tree that encompasses all of life. The end result is a digital resource that available free online for anyone to use or edit, much like a “Wikipedia” for evolutionary trees.
“This is the first real attempt to connect the dots and put it all together,” said principal investigator Karen Cranston of Duke University. “Think of it as Version 1.0.”
The current version of the tree -- along with the underlying data and source code -- is available to browse and download at tree.opentreeo...flife.org.
It is also described in an article appearing Sept. 18 in the Proceedings of the National Academy of Sciences.
Evolutionary trees, branching diagrams that often look like a cross between a candelabra and a subway map, aren’t just for figuring out whether aardvarks are more closely related to moles or manatees, or pinpointing a slime mold’s closest cousins. Understanding how the millions of species on Earth are related to one another helps scientists discover new drugs, increase crop and livestock yields, and trace the origins and spread of infectious diseases such as HIV, Ebola and influenza.
Rather than build the tree of life from scratch, the researchers pieced it together by compiling thousands of smaller chunks that had already been published online and merging them together into a gigantic “supertree” that encompasses all named species.
The initial draft is based on nearly 500 smaller trees from previously published studies.
To map trees from different sources to the branches and twigs of a single supertree, one of the biggest challenges was simply accounting for the name changes, alternate names, common misspellings and abbreviations for each species. The eastern red bat, for example, is often listed under two scientific names, Lasiurus borealis and Nycteris borealis. Spiny anteaters once shared their scientific name with a group of moray eels.
“Although a massive undertaking in its own right, this draft tree of life represents only a first step,” the researchers wrote.
For one, only a tiny fraction of published trees are digitally available.
A survey of more than 7,500 phylogenetic studies published between 2000 and 2012 in more than 100 journals found that only one out of six studies had deposited their data in a digital, downloadable format that the researchers could use.
The vast majority of evolutionary trees are published as PDFs and other image files that are impossible to enter into a database or merge with other trees.
“There’s a pretty big gap between the sum of what scientists know about how living things are related, and what’s actually available digitally,” Cranston said.
As a result, the relationships depicted in some parts of the tree, such as the branches representing the pea and sunflower families, don’t always agree with expert opinion.
Other parts of the tree, particularly insects and microbes, remain elusive.
That’s because even the most popular online archive of raw genetic sequences -- from which many evolutionary trees are built -- contains DNA data for less than five percent of the tens of millions species estimated to exist on Earth.
“As important as showing what we do know about relationships, this first tree of life is also important in revealing what we don’t know,” said co-author Douglas Soltis of the University of Florida.
To help fill in the gaps, the team is also developing software that will enable researchers to log on and update and revise the tree as new data come in for the millions of species still being named or discovered.
“It’s by no means finished,” Cranston said. “It’s critically important to share data for already-published and newly-published work if we want to improve the tree.”
“Twenty five years ago people said this goal of huge trees was impossible,” Soltis said. “The Open Tree of Life is an important starting point that other investigators can now refine and improve for decades to come.”
This research was supported by a three-year, $5.76 million grant from the U.S. National Science Foundation (1208809).
Other study co-authors include Cody Hinchliff and Stephen Smith of the University of Michigan; James Allman of Interrobang Corporation; Gordon Burleigh, Ruchi Chaudhary and Jiabin Deng of the University of Florida; Lyndon Coghill, Peter Midford and Richard Ree of the Field Museum of Natural History; Keith Crandall and Christopher Owen of George Washington University; Bryan Drew of the University of Nebraska-Kearney; Romina Gazis and David Hibbett of Clark University; Karl Gude of Michigan State University; Laura Katz and H. Dail Laughinghouse IV of Smith College; Emily Jane McTavish of the University of Kansas; Jonathan Rees of the National Evolutionary Synthesis Center and Tiffani Williams at Texas A&M University.
The project is open source - you can see the "tree" development here: tree.opentreeoflife.org...@1
I've made a request to be updated when new information or research is available.
Also, the ATS library has been extensively updated with new pages and a lot of videos. If anyone has any suggestions for additional articles or topics, please post in the sticky thread.
I am changing the GENOMICS topic to SPECIATION as I think that subject generates a lot of questions and includes genomics.
TREE OF LIFE PROJECT
Contributed by SuperFrog
"Tree of Life" for 2.3 Million Species Released
Large, open-access resource aims to be “Wikipedia” for evolutionary history
“Twenty five years ago people said this goal of huge trees was impossible,” Soltis said. “The Open Tree of Life is an important starting point that other investigators can now refine and improve for decades to come.”
September 18, 2015 | Robin A. Smith
Durham, NC - A first draft of the “tree of life” for the roughly 2.3 million named species of animals, plants, fungi and microbes -- from platypuses to puffballs -- has been released.
A collaborative effort among eleven institutions, the tree depicts the relationships among living things as they diverged from one another over time, tracing back to the beginning of life on Earth more than 3.5 billion years ago.

Tens of thousands of smaller trees have been published over the years for select branches of the tree of life -- some containing upwards of 100,000 species -- but this is the first time those results have been combined into a single tree that encompasses all of life. The end result is a digital resource that available free online for anyone to use or edit, much like a “Wikipedia” for evolutionary trees.
“This is the first real attempt to connect the dots and put it all together,” said principal investigator Karen Cranston of Duke University. “Think of it as Version 1.0.”
The current version of the tree -- along with the underlying data and source code -- is available to browse and download at tree.opentreeo...flife.org.
It is also described in an article appearing Sept. 18 in the Proceedings of the National Academy of Sciences.
Evolutionary trees, branching diagrams that often look like a cross between a candelabra and a subway map, aren’t just for figuring out whether aardvarks are more closely related to moles or manatees, or pinpointing a slime mold’s closest cousins. Understanding how the millions of species on Earth are related to one another helps scientists discover new drugs, increase crop and livestock yields, and trace the origins and spread of infectious diseases such as HIV, Ebola and influenza.
Rather than build the tree of life from scratch, the researchers pieced it together by compiling thousands of smaller chunks that had already been published online and merging them together into a gigantic “supertree” that encompasses all named species.
The initial draft is based on nearly 500 smaller trees from previously published studies.
To map trees from different sources to the branches and twigs of a single supertree, one of the biggest challenges was simply accounting for the name changes, alternate names, common misspellings and abbreviations for each species. The eastern red bat, for example, is often listed under two scientific names, Lasiurus borealis and Nycteris borealis. Spiny anteaters once shared their scientific name with a group of moray eels.
“Although a massive undertaking in its own right, this draft tree of life represents only a first step,” the researchers wrote.
For one, only a tiny fraction of published trees are digitally available.
A survey of more than 7,500 phylogenetic studies published between 2000 and 2012 in more than 100 journals found that only one out of six studies had deposited their data in a digital, downloadable format that the researchers could use.
The vast majority of evolutionary trees are published as PDFs and other image files that are impossible to enter into a database or merge with other trees.
“There’s a pretty big gap between the sum of what scientists know about how living things are related, and what’s actually available digitally,” Cranston said.
As a result, the relationships depicted in some parts of the tree, such as the branches representing the pea and sunflower families, don’t always agree with expert opinion.
Other parts of the tree, particularly insects and microbes, remain elusive.
That’s because even the most popular online archive of raw genetic sequences -- from which many evolutionary trees are built -- contains DNA data for less than five percent of the tens of millions species estimated to exist on Earth.
“As important as showing what we do know about relationships, this first tree of life is also important in revealing what we don’t know,” said co-author Douglas Soltis of the University of Florida.
To help fill in the gaps, the team is also developing software that will enable researchers to log on and update and revise the tree as new data come in for the millions of species still being named or discovered.
“It’s by no means finished,” Cranston said. “It’s critically important to share data for already-published and newly-published work if we want to improve the tree.”
“Twenty five years ago people said this goal of huge trees was impossible,” Soltis said. “The Open Tree of Life is an important starting point that other investigators can now refine and improve for decades to come.”
This research was supported by a three-year, $5.76 million grant from the U.S. National Science Foundation (1208809).
Other study co-authors include Cody Hinchliff and Stephen Smith of the University of Michigan; James Allman of Interrobang Corporation; Gordon Burleigh, Ruchi Chaudhary and Jiabin Deng of the University of Florida; Lyndon Coghill, Peter Midford and Richard Ree of the Field Museum of Natural History; Keith Crandall and Christopher Owen of George Washington University; Bryan Drew of the University of Nebraska-Kearney; Romina Gazis and David Hibbett of Clark University; Karl Gude of Michigan State University; Laura Katz and H. Dail Laughinghouse IV of Smith College; Emily Jane McTavish of the University of Kansas; Jonathan Rees of the National Evolutionary Synthesis Center and Tiffani Williams at Texas A&M University.
edit on 30-9-2015 by Phantom423 because: (no reason given)
edit on 30-9-2015 by Phantom423 because: (no reason
given)
edit on 30-9-2015 by Phantom423 because: (no reason given)
edit on 9/30/2015 by semperfortis because: Corrected
all CAPS
a reply to: Phantom423
This is amazing. Thanks for posting it here. Understandably, it is not 100% accurate or anywhere near complete, but it is a wonderful conglomeration of thousands of peoples work. It won't take long to refine this model now that so many pieces of the puzzle are on the same table. When they start including genetic info, it will most likely be a paradigm shift in the way we think of speciation, taxonomy, and evolution as a whole.
This is amazing. Thanks for posting it here. Understandably, it is not 100% accurate or anywhere near complete, but it is a wonderful conglomeration of thousands of peoples work. It won't take long to refine this model now that so many pieces of the puzzle are on the same table. When they start including genetic info, it will most likely be a paradigm shift in the way we think of speciation, taxonomy, and evolution as a whole.
originally posted by: Woodcarver
a reply to: Phantom423
This is amazing. Thanks for posting it here. Understandably, it is not 100% accurate or anywhere near complete, but it is a wonderful conglomeration of thousands of peoples work. It won't take long to refine this model now that so many pieces of the puzzle are on the same table. When they start including genetic info, it will most likely be a paradigm shift in the way we think of speciation, taxonomy, and evolution as a whole.
Yes, the open source material is fascinating. This type of project will go on for years - maybe forever because as we move out into the Solar system i.e. Mars, what if we find life that's similarly constructed? The question would become - is the DNA code ubiquitous?
I agree about the genetic links - imagine when we have the genetic blueprint for all the organisms in the tree - and then link the relationships.
edit on 30-9-2015 by Phantom423 because: (no reason given)
new topics
-
BIDEN Admin Begins Planning For January 2025 Transition to a New President - Today is 4.26.2024.
2024 Elections: 27 minutes ago -
Big Storms
Fragile Earth: 1 hours ago -
Where should Trump hold his next rally
2024 Elections: 4 hours ago -
Shocking Number of Voters are Open to Committing Election Fraud
US Political Madness: 5 hours ago -
Gov Kristi Noem Shot and Killed "Less Than Worthless Dog" and a 'Smelly Goat
2024 Elections: 6 hours ago -
Falkville Robot-Man
Aliens and UFOs: 6 hours ago -
James O’Keefe: I have evidence that exposes the CIA, and it’s on camera.
Whistle Blowers and Leaked Documents: 7 hours ago -
Australian PM says the quiet part out loud - "free speech is a threat to democratic dicourse"...?!
New World Order: 7 hours ago -
Ireland VS Globalists
Social Issues and Civil Unrest: 8 hours ago -
Biden "Happy To Debate Trump"
2024 Elections: 8 hours ago
5
